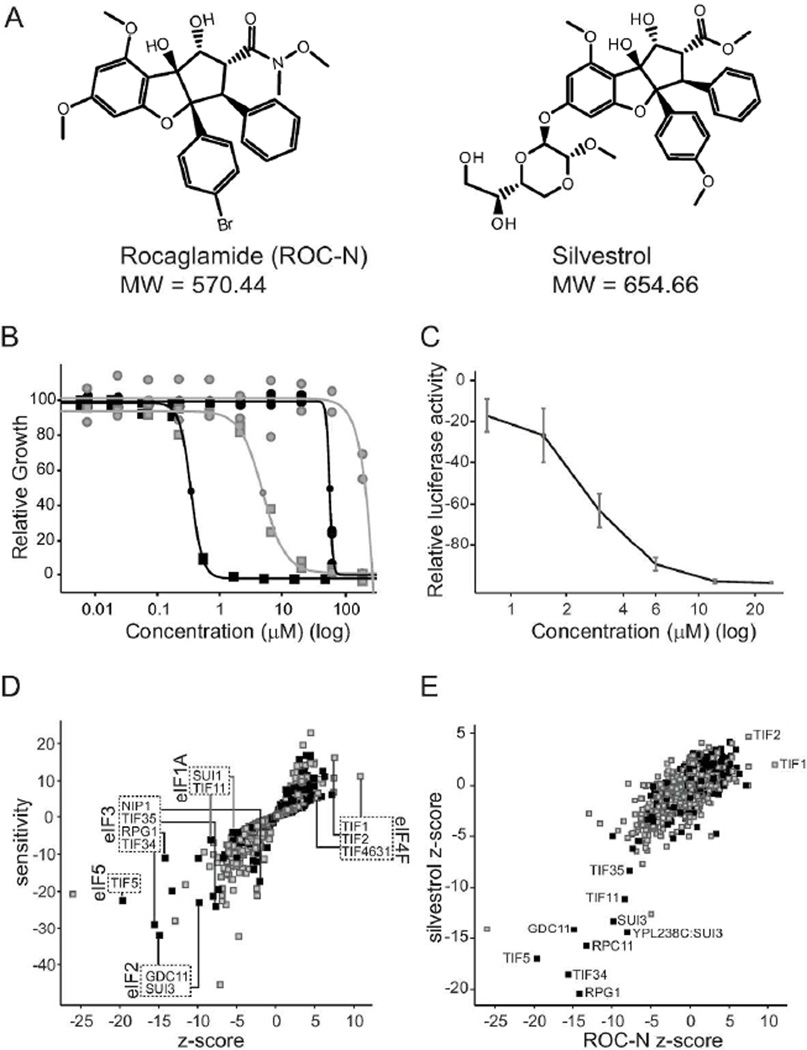
Figure 1.
Components of the translation initiation pathway alter the cellular response to rocaglamides. A. Chemical scaffolds examined in this study. B. Normalized growth (measured by OD600) of wild-type (gray) and Δ7 (black) yeast strains in the presence of ROC-N (squares) and silvestrol (circles). Shown are biological and technical replicates. n = 2. C. ROC-N inhibits expression of a luciferase reporter. Luciferase levels relative to a DMSO control are plotted versus compound concentration (log). n=3 ± SEM D. Haploinsufficiency profile of ROC-N (6 µM) with relative strain sensitivity plotted as a function of statistical significance (z-score). Strains essential for viability (black) and non-essential (gray) are indicated. Deletion strains involved in key translation initiation complexes are highlighted, together with the corresponding mammalian nomenclature annotated. E. Haploinsufficiency profile (z-score) of ROC-N (6 µM) is plotted as a function of silvestrol (200 µM). Commonly affected strains are highlighted.