Abstract
Lung cancer remains the leading cause of cancer-related deaths worldwide and non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer. With a variety of biological functions, Prohibitin1 (PHB1) has been proved tumor-associated. But there are conflicting data regarding the involvement of PHB1 in tumorigenesis and few studies regarding the role of PHB1 in lung cancer. The studies reported herein used a combination of clinical observations and molecular methods to investigate the possible role of PHB1 in NSCLC tissues and cell lines. PHB1 expression was evaluated by RT-PCR, RT-qPCR, Western blotting and immunohistochemistry analysis. Flow cytometric analysis was used to determine the surface expression profiles of PHB1 in lung cell lines. The results showed that PHB1 expression were generally increased in lung cancer tissues when compared with matched noncancerous tissues and closely related with tumor differentiation and lymph node invasion. PHB1 expression levels was also increased in three lung cancer cell lines (SK-MES-1, NCI-H157 and NCI-H292) as compared with BEAS-2B cells. Moreover, there were various subcellular localization of PHB1 in different lung cancer cells and the presence of PHB1 on the surface of lung cancer cells was significantly reduced. In conclusion, PHB1 expression is increased in NSCLC and the up-regulation of PHB1 is associated with clinically aggressive phenotype. The different subcellular localization of PHB1 in NSCLC cells and the loss of the membrane-associated PHB1 probably related to the tumorigenesis and progression of NSCLC and suggests that PHB1 may play different roles in various types of NSCLC.
Keywords: Prohibitin 1, up-regulation, subcellular localization, non-small cell lung cancer
Introduction
Prohibitin 1 (PHB1), a highly conserved ubiquitous eukaryotic protein, is belong to prohibitin family and encoded by nuclear genes PHB which is located on chromosome 17q21 [1]. Alteration of PHB1 levels has been associated with cancer [2,3]. But there are conflicting data regarding the involvement of PHB1 in tumorigenesis. PHB1 has been shown have a action to block cell cycle progression [4] and could inhibits the androgen-dependent growth of prostate cells [5], but has also been found to be overexpressed in many tumors such as chemical-induced carcinoma, hepatocellular carcinomas, some cancer cells lines and primary tumor samples [6,7]. Such opposing results regarding the role of PHB1 in tumorigenesis may be explained by its subcellular localization. Indeed, the subcellular localization of PHB1 has been shown to affect cell fate [8]. PHB1 have been found to be localized to several cellular compartments such as mitochondria, plasma membrane and nucleus, and have been implicated in the stabilization of mitochondrial proteins, transcriptional regulation, the regulation of sister chromatid cohesion, and cellular signaling [1,9,23]. Although many studies have reported that PHB1 is increased in many types of tumors, only a few reports describe its subcellular localization.
Lung cancer remains the leading cause of cancer-related deaths worldwide, being greater than that of breast, prostate and colon combined [10], and the morbidity and mortality of lung cancer is increasing recent years in China [11]. Although PHB1 is proved tumor-associated, but knowledge about the function of PHB1 in cancer is limited and there are few studies regarding the role of PHB1 in lung carcinoma.
Combined with clinical feature, this research purposed to discuss the role of PHB1 in non-small cell lung cancer (NSCLC) and the different subcellular localization of PHB1 in lung cancer cell lines.
Materials and methods
Patients and tissue samples
Ethical approval for this study was obtained by the Ethical Committee of Kunming Institute of Zoology, the Chinese Academy of Sciences. 55 surgical removal samples of NSCLC cancer tissues and peritumoral normal lung tissues were collected and confirmed by pathological assessment in the First Affiliated Hospital of Kunming Medical College between January 2009 and January 2012. Immediately after removal, all tissues for molecular analysis were put in liquid nitrogen and stored at -80°C until use. Cancer tissues for IHC were fixed in 10% neutral formalin and desiccated and embedded in paraffin.
Cell culture
Human lung epithelial cells of BEAS-2B (normal human bronchial epithelial cell line), SK-MES-1 (human lung squamous carcinoma cell line), NCI-H157 (human lung adenocarcinoma cell line) and NCI-H292 (human lung squamous carcinoma cell line - lymph node metastatic) were obtained from the American Type Culture Collection (Manassas, VA, USA). BEAS-2B cells were grown in GIBCO® LHC serum-free media. SK-MES-1, NCI-H157 were cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 containing 10% fetal calf serum (FCS). NCI-H292 were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. These cells were grown on plastic culture plates at 37°C under a humidified atmosphere of 95% air and 5% CO2 (v/v). Cultured cells were harvested at 90-95% confluence before analysis.
RNA extraction and polymerase chain reaction (PCR)
Total RNA of tissues were extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) with DNase I treatment. Total cellular RNA of the cell lines were isolated using the TRI Reagent (Molecular Research Center, Inc., USA) following the manufacturer’s protocol. The first-strand cDNA synthesis were performed as previously described [12]. For semi-quantitative reverse-transcribed PCR (RT-PCR), the primers (forward and reverse) used were as follows: PHB1: Forward: 5’-GTGGTTGGGGAATTCATGTGGAGGT-3’, Reverse: 5’-GGACGGCAGCACACGCTCAT-3’ (429 bp product); glyceraldehyde 3-phosphate dehydrogenase (GAPDH): Forward: 5’-TCGGAGTCAACGGATTTGGTCGTA-3’ and Reverse: 5’- AGCCTTCTCCATGGTGGTGAAGA-3’ (321 bp product). Amplicons were separated by electrophoresis in 2% agarose gel, stained with ethidium bromide and viewed under ultraviolet illumination. Real-time quantitative PCR (RT-qPCR) was performed with a continuous fluorescence detector (Opticon Monitor, Bio-Rad, IQ5, Hercules, CA, USA). PCR reaction was carried out using an SYBR Green real-time PCR kit (TaKaRa, Dalian, China) with the condition as the following: initial denaturation at 95°C for 1 min, followed by 40 cycles at 95°C for 15 s, 60°C for 15 s, and 76°C for 20 s. Oligonucleotide primers used were forward 5’-GCAGGACATTGTGGTAGGGG-3’ and reverse 5’-GCTGGTGAAGATGCGAGGAA-3’ for PHB1 (193 bp product); and forward 5’-ATGGGGAAGGTGAAGGTCG-3’, reverse 5’-GGGGTCATTGATGGCAACAATA-3’ for GAPDH (107 bp product) as internal standard. The identity of the RT-PCR and RT-qPCR products were confirmed by DNA sequencing. RT-qPCR for each gene was performed in triplicates in at least three separate amplifications. No template controls (no cDNA in PCR) were run to detect unspecific or genomic amplification and primer dimerization. Signal intensity of PHB1 was then quantitated by PDQuest software (Bio-Rad; Hercules, CA) and normalized to GAPDH signal intensity. Fluorescence curve analysis was carried using Opticon Monitor software. Relative quantitative evaluation of PHB1 levels were performed by E-method and expressed as a ratio of the transcript of PHB1 to GAPDH in the tumor tissue divided by a similar ratio in the non-neoplastic tissue of the same patient.
Tissue immunohistochemistry
Five micron-thick sections were obtained from Paraffin blocks of Lung tissues, dewaxed in xylene, dehydrated in ethanol, washed 3 times with distilled water, and then treated for 5 minutes in 10 mmol/L citrate buffer solution (pH 6) in an autoclave at 121°C, cooled at room temperature and rinsed in phosphate-buffered saline (PBS). Sections were pre-incubated with 2% normal rabbit serum to block nonspecific binding of antibodies for 30 minutes and then incubated overnight with the mouse monoclonal anti-human PHB1 antibody (Clone II-14-10, 1:100, NeoMarkers For Lab Vision Corporation, Thermo Fisher Scientific CA, USA) at 4°C. Specific binding was detected by a streptavidin-biotin-peroxidase assay kit (Maxim, Fujian, China). The section was counterstained with Harris hematoxylin. Direct microscopic micrographs were captured using a Leica DFC320 camera controlled by Leica IM50 software (Leica, Germany). Sections incubated with normal rabbit IgG were served as a negative control. Immunohistochemical staining was assessed semi-quantitatively by measuring both the intensity of the staining (0, 1, 2, or 3) and extent of staining (0, 0%; 1, 0-10%; 2, 10-50%; 3, 50-100%). The scores for the intensity and extent of staining were multiplied to give a weighted score for each case (maximum possible, 9). For the statistical analysis, the weighted scores were grouped into two categories where scores of 0-3 were considered negative and 4-9 positive [13].
Protein extraction
Frozen lung tissues were pulverized in liquid nitrogen using mortar and pestle. The resulting powder was solubilized in Radio Immunoprecipitation Assay (RIPA) lysis buffer (P0013C, Beyotime, China). Cultured cells were harvested at 90-95% confluence, and washed twice with ice-cold PBS. Total cellular proteins were extracted by lysing cells in the same urea lysis buffer for 60 min on ice. Soluble proteins were recovered after centrifugation at 4°C for 20 min at 12,000 ×g.
The nuclear and mitochondrial proteins of cultured cells were extracted using the Cell Mitochondria Isolation Kit (C3601, Beyotime, China) and Nuclear and Cytoplasmic Protein Extraction Kit (P0027, Beyotime, China) following the manufacturer’s protocol respectively. The protein concentration was determined by a protein assay kit (Bio-Rad, Hercules, CA, USA), and then diluted to the same concentration (5 mg/ml) with lysis buffer and stored at -80°C until future use.
Western blotting
The methods used were mainly as previously described [28]. Briefly, protein samples (containing 50 μg of protein) of tissue and cells were separated on a 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then electro-transferred onto a PVDF membrane (Millipore), subsequently blocked with 3% bovine serum albumin (BSA) for 2 h prior overnight incubation at 4°C with PHB1 monoclonal antibody (Clone II-14-10, 1:500, NeoMarkers For Lab Vision Corporation, Thermo Fisher Scientific CA, USA). The membrane were washed with 0.1% TBST (50 mM Tris pH 8.0, 150 mM NaCl, and 0.1% Tween-20 (v/v)), and then incubated with horseradish peroxidase conjugated secondary antibody for 2 h. Protein bands were visualized with Super Signal reagents (Pierce, Rockford, IL, USA) and exposure to Kodak film.
Flow cytometric analysis
To detect surface expression of PHB1 by immunofluorescence, cells were fixed with 1% paraformaldehyde in PBS at room temperature for 30 min and then blocked with 5% BSA in PBS for 30 min at 37°C. The cells were washed and then incubated with the primary antibody (anti-PHB1 antibody, 1:100) at room temperature, 1 h. PBS washed three times and incubated with secondary antibody (FITC-donkey anti goat, 1:100) at room temperature 30 min. After being washed three times with PBS, the sample was analyzed with a flow cytometer (FACS Vantage SE, Becton Dickinson, Franklin Lakes, NJ, USA) [28].
Statistical analysis
All statistical analyses were performed by the SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The Fisher’s exact test and chi squared test for the significance of correlations between PHB1 expression and clinicopathological parameters (Tables 1 and 2). Differences in the numerical data between the cancer and peritumoral normal tissues were evaluated using the paired Wilcoxon test (Figure 1B). A value of p < 0.05 was considered as significantly different.
Table 1.
Association between the mRNA levels of PHB1 and clinic-pathologic data in NSCLC patients
| Total (n = 55) | PHB1 mRNA levels | P | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Increased* (n = 37) | Not increased* (n = 18) | |||||
|
| ||||||
| No. | No. | % | No. | % | ||
| Gender | ||||||
| Male | 39 | 27 | 69.2 | 12 | 30.8 | 0.629 |
| Female | 16 | 10 | 62.5 | 6 | 37.5 | |
| Age | ||||||
| ≤ 60 | 37 | 24 | 64.9 | 13 | 35.1 | 0.585 |
| > 60 | 18 | 13 | 72.2 | 5 | 27.8 | |
| Subtype | ||||||
| Squamous-celled carcinoma | 16 | 9 | 56.3 | 7 | 43.8 | 0.302 |
| Adenocarcinoma | 34 | 25 | 73.5 | 9 | 26.5 | |
| Adenosquamous carcinoma | 5 | 3 | 60.0 | 2 | 40.0 | |
| Differentiation | ||||||
| Well | 21 | 10 | 47.6 | 11 | 52.4 | 0.015 |
| Poor and moderated | 34 | 27 | 79.4 | 7 | 20.6 | |
| Lymph node metastasis | ||||||
| Positive | 28 | 24 | 85.7 | 4 | 14.3 | 0.003 |
| Negative | 27 | 13 | 48.1 | 14 | 51.9 | |
| Distant metastasis | ||||||
| Positive | 6 | 4 | 66.7 | 2 | 33.3 | 0.973 |
| Negative | 49 | 33 | 67.3 | 16 | 32.7 | |
| Tumor size | ||||||
| ≤ 3 cm | 25 | 16 | 64.0 | 9 | 36.0 | 0.637 |
| > 3 cm | 30 | 21 | 70.0 | 9 | 30.0 | |
The increased folds of “> 2” were defined as “increased”, while the increased folds of “≤ 2” were defined as “not increased”.
Table 2.
Association between the protein levels of PHB1 with histopathologic features of NSCLC patients
| Total (n = 26) | PHB1 expression | P | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Increased (n = 22) | Not Increased (n = 4) | |||||
|
| ||||||
| No. | No. | % | No. | % | ||
| Gender | ||||||
| Male | 19 | 15 | 78.9 | 4 | 21.1 | 0.187 |
| Female | 7 | 7 | 100.0 | 0 | 0.0 | |
| Age | ||||||
| ≤ 60 | 19 | 16 | 84.2 | 3 | 15.8 | 0.925 |
| > 60 | 7 | 6 | 85.7 | 1 | 14.3 | |
| Subtype | ||||||
| Squamous-celled carcinoma | 15 | 12 | 80.0 | 3 | 20.0 | 0.188 |
| Adenocarcinoma | 10 | 9 | 90.0 | 1 | 10.0 | |
| Adenosquamous carcinoma | 1 | 1 | 100.0 | 0 | 0.0 | |
| Differentiation | ||||||
| Well | 8 | 5 | 62.5 | 3 | 37.5 | 0.037 |
| Poor and moderated | 18 | 17 | 94.4 | 1 | 5.6 | |
| Lymph node metastasis | ||||||
| Positive | 16 | 15 | 93.8 | 1 | 6.3 | 0.102 |
| Negative | 10 | 7 | 70.0 | 3 | 30.0 | |
| Distant metastasis | ||||||
| Positive | 4 | 4 | 100.0 | 0 | 0.0 | 0.354 |
| Negative | 22 | 18 | 81.8 | 4 | 18.2 | |
| Tumor size | ||||||
| ≤ 3 cm | 12 | 10 | 83.3 | 2 | 16.7 | 0.867 |
| > 3 cm | 14 | 12 | 85.7 | 2 | 14.3 | |
Figure 1.
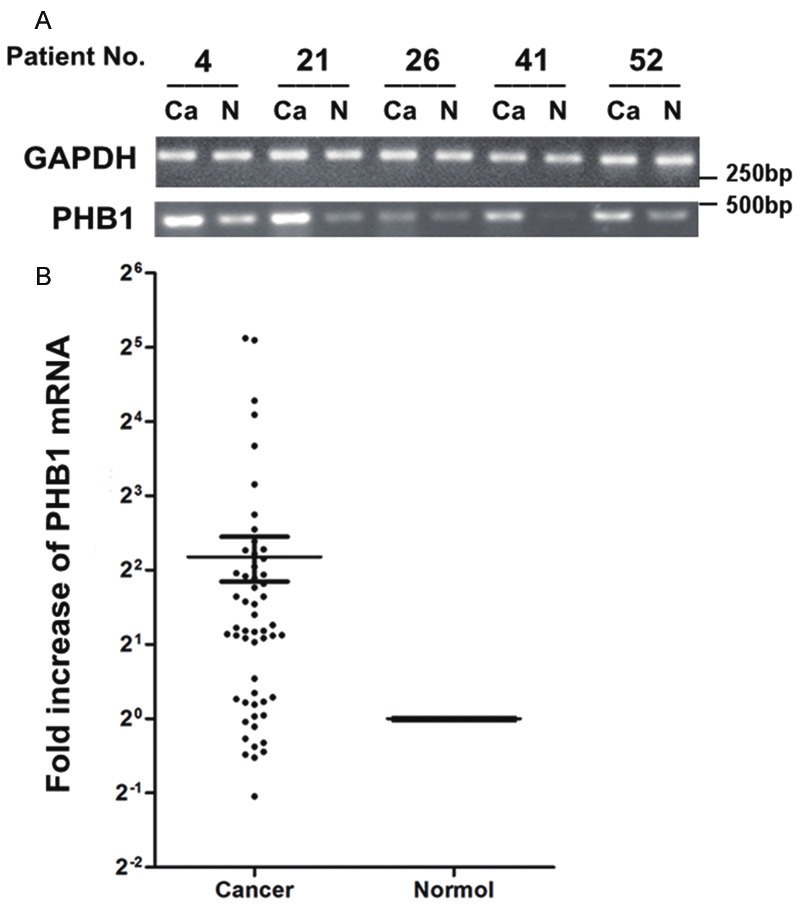
Expression of PHB1 in lung cancer tissues measured by RT-PCR and RT-qPCR. A. The matched cancerous (Ca) and relevant normal (N) tissues from each patient were analyzed by RT-PCR using GAPDH- and PHB1-specific primers (n = 5). After normalized to GAPDH levels, the results of RT-PCR showed significantly increased mRNA levels of PHB1 in cancer compared to normal. B. PHB1 expression in 55 NSCLC samples was measured by RT-qPCR. PHB1 mRNA was increased in 67.3% (37 of 55) tumors. Mean fold increase in the tumor tissue relative to non-neoplastic lung tissue was shown. Bars, SEM. The increased folds of “> 2” were defined as “increased”, whereas increased folds of “≤ 2” were defined as “not increased”.
Results
PHB1 mRNA was up-regulated frequently in lung NSCLC and correlated with clinicopathological parameters
RT-PCR was performed on matched normal and cancer tissue specimens from 5 select randomly patients after the samples were normalized to GAPDH levels, and the results showed significant increased mRNA levels of PHB1 in cancer tissues compared to normal tissues (Figure 1A). The differential expression of PHB1 mRNA was further examined by RT-qPCR analysis in 55 NSCLC samples. Overall up-regulated PHB1 expression was detected in 67.3% (37 of 55) of NSCLC tissues when compared the primary tumor with matched non-neoplastic tissue (Figure 1B). PHB1 mRNA was increased 4.87 ± 1.1 fold (mean ± SEM) in tumors (p = 0.0198, paired Wilcoxon test). Furthermore, there were significant differences in PHB1 mRNA expression in well versus poor and moderated differentiation tumors (p = 0.015, chi squared test), and lymphonode metastasis versus non-metastasis tumors (p = 0.003) (Table 1). There have no obvious difference between subtypes (p = 0.302).
Protein levels of PHB1 are frequently increased in the cancerous lung tissues and are correlated with histopathologic features
The presence of PHB1 in the normal lung tissues was verified using Western blotting (Figure 2A). However, in patients with NSCLC, a marked up-regulated of PHB1 expression was observed in all cases (4/4) NSCLC tissues comparing the matched nonmalignant lung tissue (Figure 2A). In immunohistochemical assay, we found that PHB1 was weakly expressed in non-neoplastic lung epithelial cells, but the staining level was significantly increased in NSCLC tissues, especially in the metastatic carcinoma tissues within lymph node (Figure 2B). PHB1 staining was mainly diffused throughout the cytoplasm and membrane of cancer cells. Overall PHB1 expression was increased in 84.6% (22/26) of NSCLC tissues (Table 2). Immunohistochemical staining of PHB1 in NSCLC tissues was graded and classified by criteria described in Section 2. We further investigated the correlation of clinical significance between up-regulated of PHB1 expression and clinicopathological parameters (Table 2). Statistical analysis showed that increased PHB1 expression was strongly associated with tumor differentiation. PHB1 expression was increased in 62.5% of well differentiated cancers and 94.4% of poor and moderated differentiated cancers (p = 0.037, chi squared test) (Table 2). There have no obvious difference between subtypes of NSCLC (p = 0.188).
Figure 2.
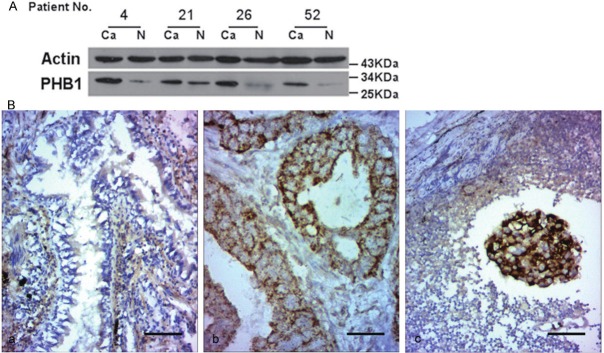
Western blotting and immunohistochemistry analysis of PHB1 in normal and cancerous lung tissues. A. Western blotting of tissue lysates from 4 cases of cancer (Ca) and relevant normal tissues (N). The expression of actin was served as a control. The significant increase of PHB1 was observed in the cancerous tissues in contrast to their matched normal tissues. B. Representative photomicrographs of immunohistochemical staining for PHB1 expression in (a) normal lung; (b) lung adenocarcinoma; (c) lung squamous carcinoma (lymph node metastatic). PHB1 staining was mainly diffused throughout the cytoplasm and membrane of cancer cells. Bar, 50 μm for each panel.
Up-regulation of PHB1 in lung cancer cell lines and the different subcellular distribution of PHB1 protein
RT-PCR was performed on BEAS-2B and three cancer cell lines after the cells were normalized to GAPDH levels. The results showed that PHB1 mRNA expression was increased in all cancer cells when compared with BEAS-2B cells (Figure 3A). RT-qPCR results showed relative expression of PHB1 mRNA in SK-MES-1, NCI-H157, NCI-H292 were 1.7, 18.2, 25.8-fold respectively when compared with BEAS-2B (Figure 3B).
Figure 3.
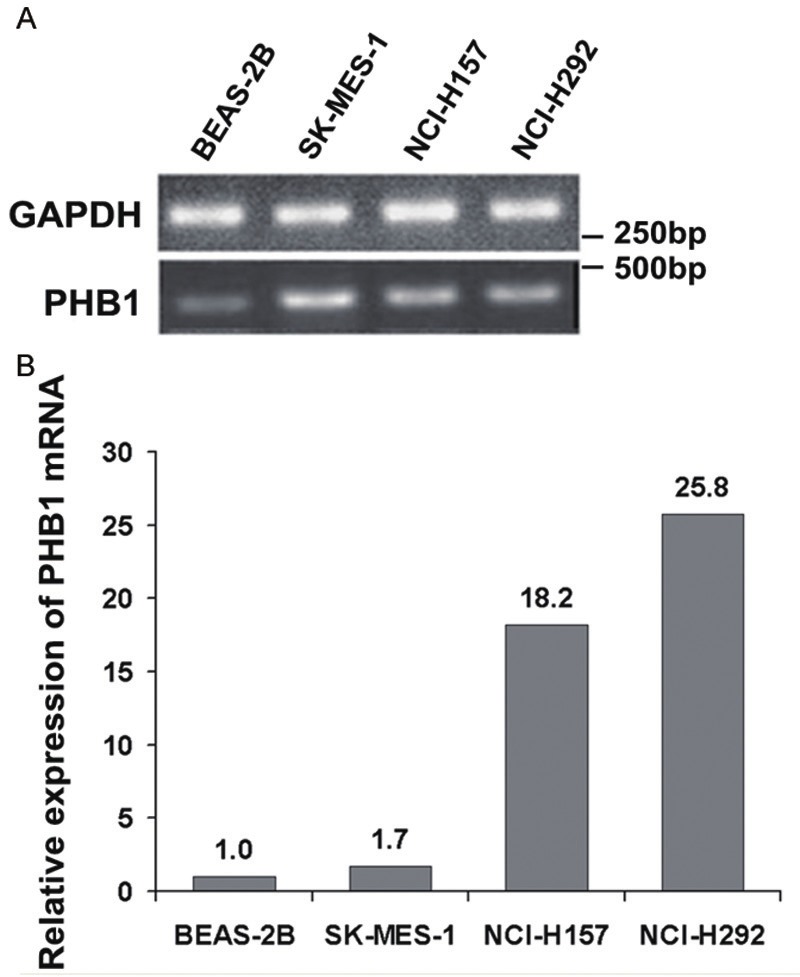
Expression of PHB1 in lung cell lines measured by RT-PCR and RT-qPCR. A. The four cell lines were analyzed by RT-PCR using GAPDH- and PHB1-specific primers. After normalized to GAPDH levels, the results of RT-PCR showed significantly increased mRNA levels of PHB1 in cancer cells compared to BEAS-2B. B. Relative expression of PHB1 mRNA in SK-MES-1, NCI-H157, NCI-H292 were 1.7, 18.2, 25.8-fold respectively when compared with BEAS-2B.
Western blot analysis revealed that PHB1 expression was significantly increased in lung cancer cell lines (Figure 4A, total protein). However, the levels of PHB1 in subcellular fractions were different. PHB1 was dominantly distributed in the mitochondrial fractions of the BEAS-2B cells. But when compared with BEAS-2B, PHB1 in nucleoprotein and mitochondrial fractions of SK-MES-1 were all increased, and in mitochondrial fractions of NCI-H157 was increased significantly but decreased in nucleoprotein. PHB1 in nucleoprotein of NCI-H292 increased obviously but decreased in mitochondrial fractions (Figure 4A). Flow cytometry with PHB1-specific antibodies was used to determine the surface expression profiles of PHB1 in the lung cancer cell lines. As shown in Figure 4B, BEAS-2B cells express relatively high levels of PHB1. NCI-H292 cells express low levels of PHB1 and little or no PHB1 was present in the SK-MES-1 and NCI-H157 cells. So the results showed that though the total level of PHB1 was up-regulated in lung cancer cells when compared with the normal cells, the cell membrane associated PHB1 was reduced or even missed.
Figure 4.
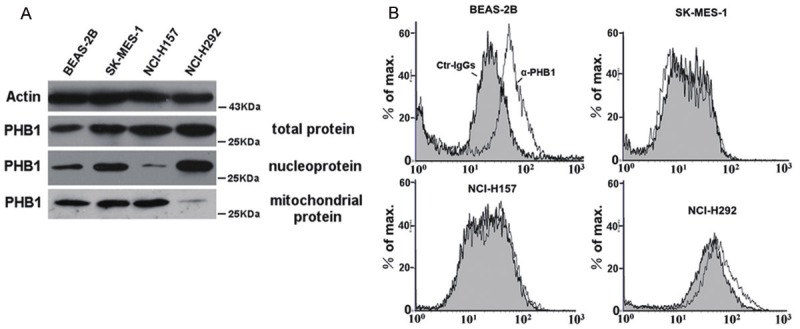
Western blotting and flow cytometric analysis for the subcellular localization of PHB1 in lung cell lines. A. Western blotting of cell lysates from four cell lines. Expression of actin was served as a control. The expression of PHB1 in total protein of all cancer cell lines were obviously increased when compared with BEAS-2B. PHB1 in nucleoprotein and mitochondrial fractions of SK-MES-1 were all increased; Mitochondrial PHB1 of NCI-H157 was increased but significantly decreased in nucleoprotein; PHB1 in nucleoprotein of NCI-H292 increased obviously but decreased in mitochondrial fractions. B. Flow cytometric analysis for surface expression of PHB1 in four lung cell lines. BEAS-2B cells express relatively high levels of PHB1. The NCI-H292 cells expresses low levels of PHB1 and little or no PHB1 was present in the SK-MES-1 and NCI-H157 cells.
Discussion
The present study showed that PHB1 is up-regulated at both the transcriptional and translational levels in NSCLC tissues when compared with normal lung tissues. The finding that overexpression of PHB1 is correlated with the malignant degree and progression of NSCLC is consistent with those of previous studies about the expression of PHB1 in lung cancer [14,15]. Although knowledge about the function of PHB1 in cancer is limited, our results clearly showed the total PHB1 up-regulation and membrane-associated PHB1 reduction in NSCLC and it might plays an important role in lung oncogenesis.
PHB1 over-expression might be due to myc-binding elements in the promoter region of PHB1, whereas c-myc oncogene is overexpressed in many cancer cells and may facilitate PHB1 expression [6,7]. Transcriptional silencing by promoter hypermethylation has emerged as one of the important mechanisms of gastric cancer development [16]. Promoter methylation is shown to be inversely correlated with mRNA levels and demethylation maybe increased gene transcription. We further analyzed the methylation status of cytosines in sites in the PHB1 promoter region of four lung epithelial cell lines, but no methylation sites were detected (data not showed).
PHB1 is mainly localized in the inner mitochondrial membrane and is essential to maintain mitochondrial homeostasis and gene expression [17]. It acting as foldase–unfoldase molecular chaperones, and therefore stabilizing the new synthesized proteins in mitochondria [18]. On the other hand, some reports have also shown that PHB1 is sometimes located in the nucleus and is involved in transcriptional regulation through interacting with several transcription factors such as E2F, p53 and steroid hormone receptors [6,19]. Phb1 is also present on the cell surface in a variety of cell types, such as B lymphocytes [20], adipose endothelial cells [21], intestinal epithelial cells [22], platelet [23] and some tumor cells [24], and have been reported to be involved in typhoid fever, obesity, platelet aggregation and cancer metastasis. But there has not been a systematic study of the subcellular localization of PHB1 in various cell types.
Our study is the first to show the different subcellular localization of PHB1 in different types of NSCLC cells. In this research, the total PHB1 levels of three lung cancer cell lines were all up-regulated when compared with BEAS-2B, but the subcellular localization of PHB1 was different and the presence of PHB1 on the surface of lung cancer cells was significantly reduced. PHB1 is differentially distributed between lung normal and cancer cells as well as in various NSCLC cells. The results might be elucidated by the conclusion that PHB1 protein could shuttle between various cellular compartments, including the mitochondria, nucleus and membrane [8,24,25].
The result that mitochondrial PHB1 of NCI-H292 was apparently down-regulated when compared with SK-MES-1 cells is according with the conclusion that cancer cells rely heavily on glycolysis instead of mitochondrial electron transport system to obtain ATP for proliferation and metastasis in the presence of adequate oxygen levels [26]. In addition, the up-regulated PHB1 in nuclei of NCI-H292 cells might involved in their ability of metastasis.
We have provided evidence that PHB1 expression levels were decreased in membrane of lung cancer cells although whole cell levels of the protein were increased. This differs from a report by Rajalingam et al. [27], who reported that increased levels of PHB1 on the cell membrane is involved in tumorigenesis through its interaction with c-Raf induced by the Ras oncogene. This difference may be cell line specific. We conclude from these results that it is might the redirection of PHB1 from the cell surface to other intracellular locale that mediates the tumorigenesis or metastasis of NSCLS. We do not at this time know whether this is due specifically to the deficiency of PHB1 on the cell surface or to its removal to intracellular sites. For some tumors, alterations in PHB1 levels or interference with PHB1 localization may become a research direction of tumor prevention and treatment.
In conclusion, PHB1 is up-regulated in NSCLC as well as related cell lines and closely associated with clinically aggressive phenotype. However, the NSCLC is also related to the loss of the membrane-associated PHB1 and its different subcellular localization. Between the two controversial results, which would be better to explain the relationship between PHB1 and NSCLC need further study. Moreover, the intracellular localization of PHB1 during the progression of normal tissue to carcinoma need further study too.
Acknowledgements
This work is supported by grants from the Chinese National Natural Science Foundation (31270835, U1132601), the National Basic Research Program of China (973 Program, 2010CB529800), and the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L03).
Disclosure of conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Kang X, Zhang L, Sun J, Ni Z, Ma Y, Chen X, Sheng X, Chen T. Prohibitin: a potential biomarker for tissue-based detection of gastric cancer. J Gastroenterol. 2008;43:618–625. doi: 10.1007/s00535-008-2208-3. [DOI] [PubMed] [Google Scholar]
- 3.Xu DH, Tang J, Li QF, Shi SL, Chen XF, Liang Y. Positional and expressive alteration of prohibitin during the induced differentiation of human hepatocarcinoma SMMC-7721 cells. World J Gastroenterol. 2008;14:5008–5014. doi: 10.3748/wjg.14.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClung JK, Danner DB, Stewart DA, Smith JR, Schneider EL, Lumpkin CK, Dell’Orco RT, Nuell MJ. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun. 1989;164:1316–1322. doi: 10.1016/0006-291x(89)91813-5. [DOI] [PubMed] [Google Scholar]
- 5.Dart DA, Spencer-Dene B, Gamble SC, Waxman J, Bevan CL. Manipulating prohibitin levels provides evidence for an in vivo role in androgen regulation of prostate tumours. Endocr Relat Cancer. 2009;16:1157–1169. doi: 10.1677/ERC-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci. 2002;59:143–155. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates PJ, Nenutil R, McGregor A, Picksley SM, Crouch DH, Hall PA, Wright EG. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp Cell Res. 2001;265:262–273. doi: 10.1006/excr.2001.5166. [DOI] [PubMed] [Google Scholar]
- 8.Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem. 2006;281:2951–2959. doi: 10.1074/jbc.M508669200. [DOI] [PubMed] [Google Scholar]
- 9.Mishra S, Murphy LC, Nyomba BL, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol Med. 2005;11:192–197. doi: 10.1016/j.molmed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Cai B. The impact of tobacco on lung health in China. Respirology. 2003;8:17–21. doi: 10.1046/j.1440-1843.2003.00433.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu SB, He YY, Zhang Y, Lee WH, Qian JQ, Lai R, Jin Y. A novel non-lens betagamma-crystallin and trefoil factor complex from amphibian skin and its functional implications. PLoS One. 2008;3:e1770. doi: 10.1371/journal.pone.0001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Yu G, Jiang P, Xiang Y, Li W, Lee W, Zhang Y. Decreased expression of protease-activated receptor 4 in human gastric cancer. Int J Biochem Cell Biol. 2011;43:1277–1283. doi: 10.1016/j.biocel.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Alfonso P, Catala M, Rico-Morales ML, Durante-Rodriguez G, Moro-Rodriguez E, Fernandez-Garcia H, Escribano JM, Alvarez-Fernandez E, Garcia-Poblete E. Proteomic analysis of lung biopsies: Differential protein expression profile between peritumoral and tumoral tissue. Proteomics. 2004;4:442–447. doi: 10.1002/pmic.200300647. [DOI] [PubMed] [Google Scholar]
- 15.Nan Y, Yang S, Tian Y, Zhang W, Zhou B, Bu L, Huo S. Analysis of the expression protein profiles of lung squamous carcinoma cell using shot-gun proteomics strategy. Med Oncol. 2009;26:215–221. doi: 10.1007/s12032-008-9109-4. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 17.Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metab. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5:714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 19.Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J Cell Mol Med. 2006;10:353–363. doi: 10.1111/j.1582-4934.2006.tb00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Kohler G, Lamers MC. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994;13:3782–3792. doi: 10.1002/j.1460-2075.1994.tb06689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10:625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 22.Sharma A, Qadri A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci U S A. 2004;101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang Y, Xiang Y, Lee W. Prohibitins are involved in protease-activated receptor 1-mediated platelet aggregation. J Thromb Haemost. 2012;10:411–418. doi: 10.1111/j.1538-7836.2011.04607.x. [DOI] [PubMed] [Google Scholar]
- 24.Patel N, Chatterjee SK, Vrbanac V, Chung I, Mu CJ, Olsen RR, Waghorne C, Zetter BR. Rescue of paclitaxel sensitivity by repression of Prohibitin1 in drug-resistant cancer cells. Proc Natl Acad Sci U S A. 2010;107:2503–2508. doi: 10.1073/pnas.0910649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Lazaro M. The warburg effect: why and how do cancer cells activate glycolysis in the presence of oxygen? Anticancer Agents Med Chem. 2008;8:305–312. doi: 10.2174/187152008783961932. [DOI] [PubMed] [Google Scholar]
- 27.Rajalingam K, Rudel T. Ras-Raf signaling needs prohibitin. Cell Cycle. 2005;4:1503–1505. doi: 10.4161/cc.4.11.2142. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Yu G, Wang Y, Xiang Y, Gao Q, Jiang P, Zhang J, Lee W, Zhang Y. Activation of protease-activated receptor 1 by a frog trefoil factor and PAR4 by human trefoil factor 2. Cell Mol Life Sci. 2011;68:3771–3780. doi: 10.1007/s00018-011-0678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


