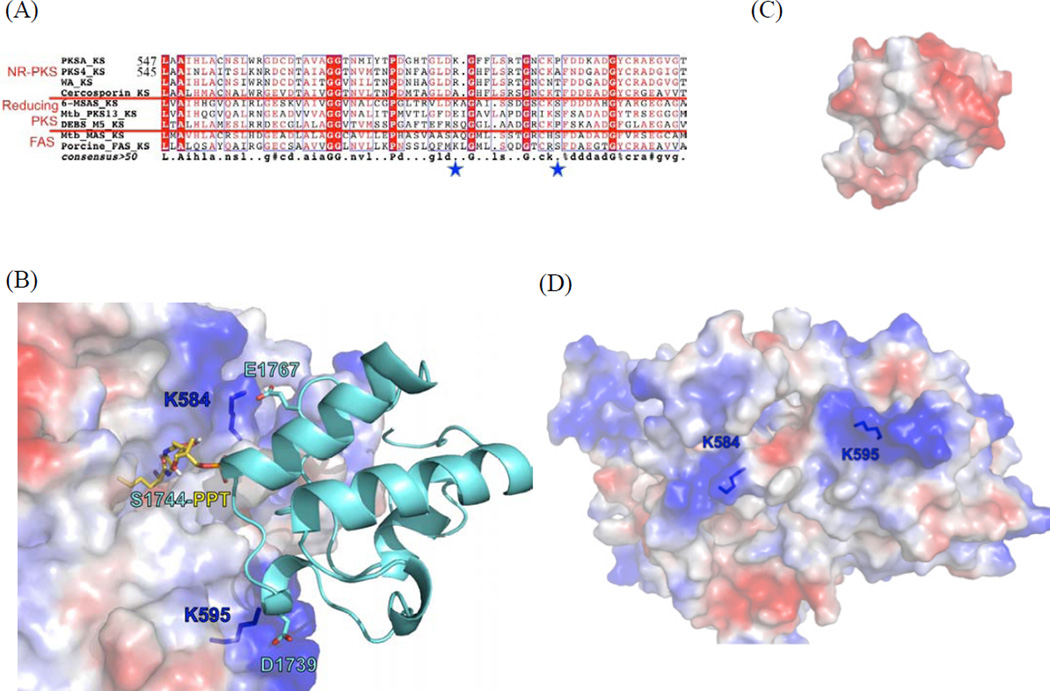
Figure 5.
see also Figure S7. Probing surface residues important for KS-ACP interactions. (A) Sequence alignment identified two relatively conserved, positively charged residues (star). The boxed residues are conserved ones. (B) Homology modeling and docking simulation of KS domains of PksA with PksA ACP suggests that K584 and K595 of PksA are located at the protein surface, where positively-charged lysines are docked with acid residues of ACP, while active site Ser tethered 2-bromohexyl pantetheine docks into the active site. D1739 and E1767 of PksA ACP were identified based on protein-protein docking simulation (Fig. S8). (C) The negatively-charged ACP surface that is docked to KS. (D) The positively-charged KS surface that is docked to ACP