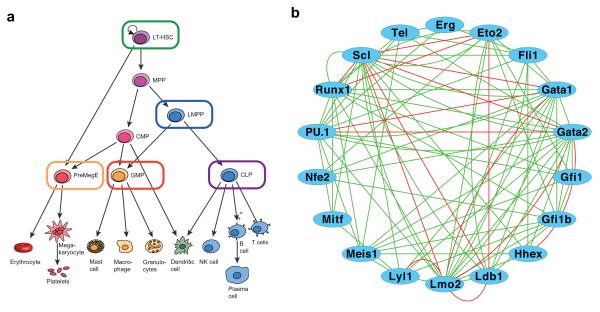
Figure 1. Single cell gene expression analysis of a core haematopoietic transcriptional regulatory network.
(a) Schematic of the haematopoietic hierarchy, with the megakaryocyte-erythroid lineage in red, the myeloid lineages in orange and the lymphoid lineage in blue. Cell types investigated in this study are outlined in the colours used to represent these populations in subsequent figures, and encompass both early multipotent stem and progenitors and committed progenitors for each of the major haematopoietic lineages. Cell surface phenotypes were LSK CD150+CD48− HSC (also gated as CD34loFlt3−), LSK Flt3hi LMPP, Lin−IL7Rα+KitloSca-1lo CLP, CD41loCD16/32hi GMP (also gated Lin−c-Kit+CD150−), CD16/32loCD41−CD150+CD105lo PreMegE (also gated Lin−c-Kit+). LT-HSC, long-term haematopoietic stem cell; MPP, multi-potent progenitor; LMPP, lymphoid-primed multi-potent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte-monocyte progenitor; PreMegE, pre megakaryocyte erythroid progenitor; NK cell, natural killer cell. (b) Network diagram of data curated from the literature and protein interaction databases (STRING66 and FunctionalNet67) illustrating the complex interactions between 18 core haematopoietic transcription factors. Green lines indicate functional relationships and red lines indicate direct protein-protein interactions. Activating and inhibitory connections are not distinguished.