Figure 2. Bootstrap to asses our results for each RNA fraction and each protein set.
Bootstrap results for total RNA fractions are shown in A (nuclear) and B (secretome). Polysomal fraction is shown in C (nuclear) and D (secretome). For each such pair of conditions, we performed a bootstrap analysis as explained in 0.6. For each miRNA we permute the values of the genes and calculate the explained variance from the resulting linear model. This procedure is repeated 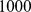 times. The y-axis represents how many times the “true” miRNA wins over the random model. The x-axis represents all miRNAs. The colors, from red to green, represent the explained variance from the current “true” model. It can be observed that the miRNAs win almost all times (the larger bars, almost reaching 1), explain the larger variance, and hence produce the best models (red).
times. The y-axis represents how many times the “true” miRNA wins over the random model. The x-axis represents all miRNAs. The colors, from red to green, represent the explained variance from the current “true” model. It can be observed that the miRNAs win almost all times (the larger bars, almost reaching 1), explain the larger variance, and hence produce the best models (red).