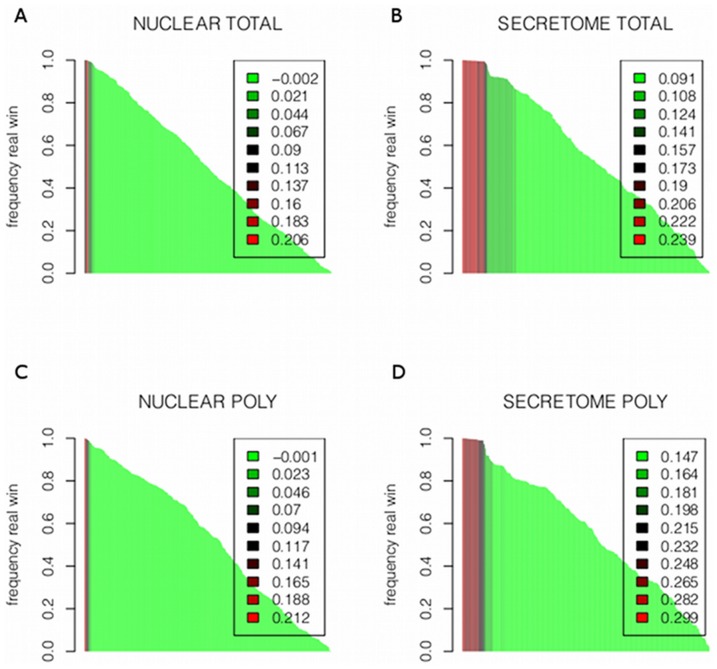
Figure 3. Linear models for day 5 secreted proteins represented graphically.
(A, B) Polysomal fraction, (C, D) total RNA. (A) and (C): plot representing logFC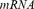 against logFC
against logFC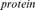 . The dashed blue line is the best fitting line of the base model,
. The dashed blue line is the best fitting line of the base model, 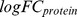 against
against 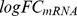 . The straight black line is the identity line (so you get an idea of the real coefficient of the model). The colored full dots are genes, which are moved after applying the model with miRNAs. Hence, they represent genes that are better explained by our model. The arrows indicate the direction of the movement. (B) and (D): plot representing our linear model including miRNA effect. In this case, the best (multivariate) model is shown: miR-130b and miR-558 (polysomal) and miR-150* (total). Full dots are the genes that were corrected by our model, being now closer to the protein prediction line of the model (red full line). Black identity line concurs with the red line. Note that the abscissas of (A) and (C) seem to have a compression of range with respect to the plots below, (B) and (D). This is not a compression, since they are different x-axis: (A) and (C) hold logFC
. The straight black line is the identity line (so you get an idea of the real coefficient of the model). The colored full dots are genes, which are moved after applying the model with miRNAs. Hence, they represent genes that are better explained by our model. The arrows indicate the direction of the movement. (B) and (D): plot representing our linear model including miRNA effect. In this case, the best (multivariate) model is shown: miR-130b and miR-558 (polysomal) and miR-150* (total). Full dots are the genes that were corrected by our model, being now closer to the protein prediction line of the model (red full line). Black identity line concurs with the red line. Note that the abscissas of (A) and (C) seem to have a compression of range with respect to the plots below, (B) and (D). This is not a compression, since they are different x-axis: (A) and (C) hold logFC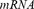 values, while (B) and (D) logFC
values, while (B) and (D) logFC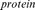 .
.