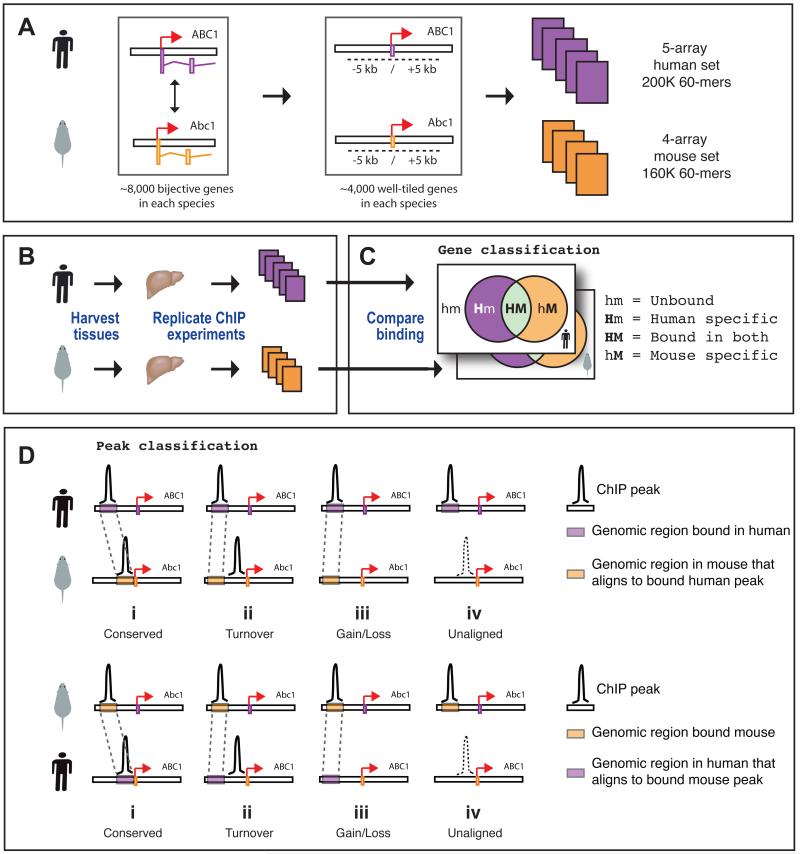
Figure 1.
Strategy to analyze transcription factor-DNA interactions in mouse and human. (A) Panel 1: approximately 8,000 high-confidence human (purple) and mouse (orange) gene orthologs were identified. Panel 2: 60-mer oligonucleotides were designed against a ten kilobase region centered around the complete set of transcription start sites in both species (colored boxes on genome track); orthologous genes with incomplete coverage, low oligonucleotide quality, or substantial gaps in one or both species were removed from the final design (Supplementary Methods). Panel 3: a human 5-array set and a mouse 4-array set capturing the transcription start sites for approximately 4,000 genes in each species were created using these oligonucleotides. (B) Mouse and human hepatocytes were isolated from liver samples and used in chromatin immunoprecipitations, which were hybridized against the array sets. (C) Gene-centric analysis classifies orthologous gene pairs by whether they are not bound in either species (hm), bound uniquely in human (Hm), bound in both species (HM), or bound uniquely in mouse (hM). (D) Peak-specific analysis classifies peaks relative to whether corresponding aligned regions exist in the second species and whether these aligned regions are bound. The four possible outcomes are shown in both the human-to-mouse and the mouse-to-human panels: In the first three cases (i, ii, iii) the aligned locus is present in the arrayed region of the ortholog. Case i (conserved): the aligned regions are bound in both species; Case ii (turnover): the orthologous gene is bound, but not at the aligned locus; Case iii (gain/loss): no binding is detected in the arrayed region of the second species, including the aligned sequence; Case iv (unaligned): the aligned sequence is not present within the arrayed region and therefore we cannot definitively classify the binding event, regardless of the presence or absence of a binding event in the other species.