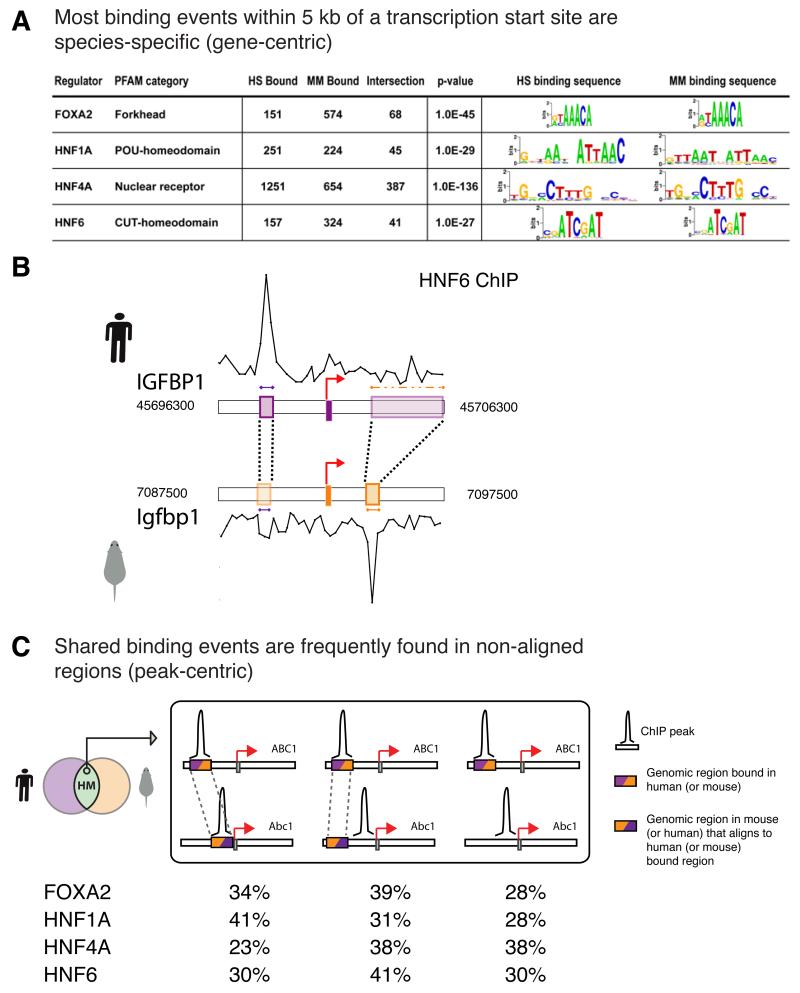
Figure 2.
(A) Number of genes bound by liver master regulators in each species, p-value (using a hypergeometric distribution) that the cross-species overlap is due to random chance, and the THEME-derived binding motifs in human and mouse. (B) The location of binding events varies between species. Here, ChIP enrichments are shown as traces. The 500 base pair sequence underlying the ChIP peak in each species is colored by species (purple human, orange mouse) and aligned with the corresponding sequence in the second species using dashed lines. For clarity, mouse ChIP enrichments are displayed as a negative y-axis, but orientation of the transcription start site is left to right. IGFPB1 is bound by HNF6 in both species, but the binding events do not align. The human sequence aligned to the mouse HNF6 peak in IGFBP1 contains large insertions overlapping a substantial portion of the human first intron (outlined with a dashed orange box), and is not bound by HNF6. (C) Shared binding events are frequently found in non-aligned regions. From left to right: aligned regions (shown as colored boxes) that are bound in both species (Figure 1D, case i); aligned regions present on both human and mouse arrays but bound only in one species (Figure 1D, case ii); regions bound in both species, but lacking aligned sequences on the orthologous array (Figure 1D, case iv, with a binding peak present). Typically, only about a third of the binding events detected in both species occur in sequences that align to each other (Table S3).