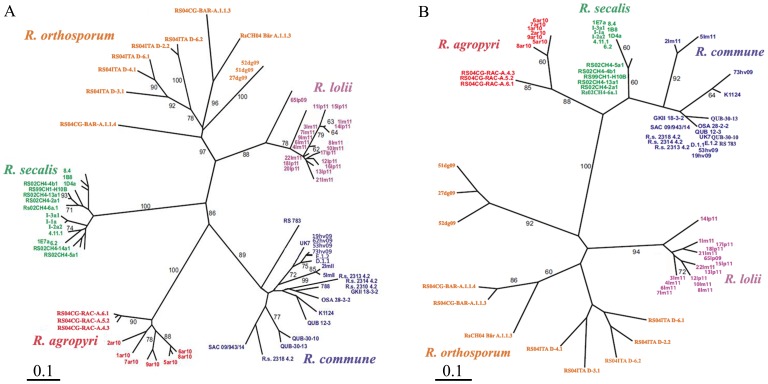
Figure 1. DNA fingerprinting methods distinguish between five Rhynchosporium species.
(A) RAPD-PCR fingerprinting of 79 isolates using combined data from seven RAPD-PCR primers; (B) rep-PCR genomic fingerprinting of 71 isolates using combined data from two primer pairs (ERIC2/BOXA1R and ERICF/BOXA1R). Both unrooted trees were constructed by neighbour-joining analyses with branch lengths drawn to show genetic distance derived from Jaccard’s coefficient of band matching (scale bar: 0.1 = 10% genetic difference). Numbers at nodes indicate the percentage bootstrap support (based on 1000 re-samplings) for the groupings; only values (A) >60% and (B) >70% are shown, for clarity. Both fingerprinting methods discriminated between isolates of R. commune (blue), R. agropyri (red), R. secalis (green), R. orthosporum (yellow) and R. lolii (purple). Note that two isolates of R. commune (2lm11 and 5lm11) were collected from Italian ryegrass.