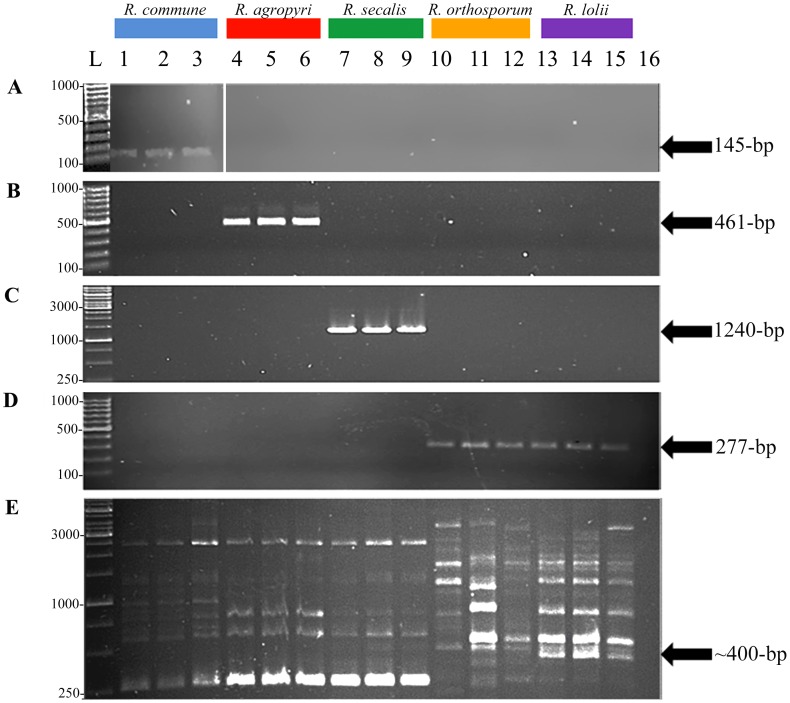
Figure 6. PCR-based diagnostic tests to distinguish between five Rhynchosporium species.
Five primer pairs (LinA-F/R; RA6-F/R; RS25-F/R; 2RO-F/R; ERIC2/BOXA1R) were tested with representative isolates of R. commune (lanes 1–3; K1124, QUB 12-3, OSA 28-2-2), R. agropyri (lanes 4–6; RS04CG-RAC-A.6.1, 6ar10, 10ar10), R. secalis (lanes 7–9; RS99CH1 H10B, E7a, I-1a), R. orthosporum (lanes 10–13; 27dg09, RS04CG-BAR-A.1.1.4, RS04ITA D-4.1) or R. lolii (lanes 13–15; 1lm11, 9lm11 and 18lp11). (A) LinA-F/R produced a 145-base pair (bp) amplicon specific for R. commune isolates; (B) RA6-F/R produced a 461-bp amplicon specific for R. agropyri isolates; (C) RS25-F/R produced a 1240-bp amplicon specific for R. secalis isolates; (D) 2ROR-F/R produced a 277-bp amplicon specific for both R. orthosporum and R. lolii isolates; use of rep-PCR genomic fingerprinting primers ERIC2/BOXA1R produced a ∼400-bp amplicon specific only for isolates of R. lolii. Different isolates are displayed in (B, C) lanes 1–3: FI12-63, QUB 9-10, 2lm11; (D) lanes 1–15∶19hv09, UK7, 2lm11, 10ar10, 6ar10, RS04CG-RAC-A.6.1, RS02CH4-4b1, RS02CH4-14a1, 6.2, 27dg09, RS04CG-BAR-A.1.1.4, RS04ITA D-4.1, 9lm11, 14lp11 and 18lp11; (E) lanes 3, 6 and 14∶2lm11, 1ar10 and 8lm11, respectively. Note that in (A) lanes 1–3 have been inserted from a different gel image. Lane labelled ‘L’ contains a 100-bp ladder (A, B, D) or a 1-kilobase ladder (C, E) (both Fermentas, UK); lane 16 is a no template control.