Figure 1.
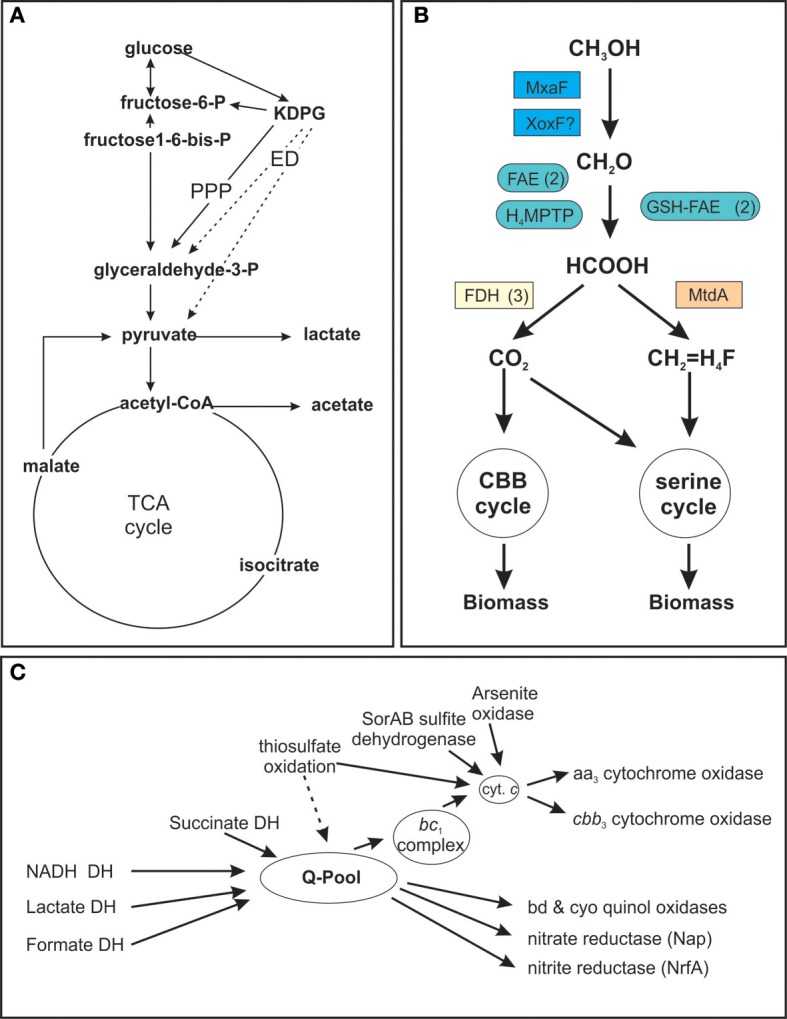
Schematic representation of S. novella metabolic pathways. (A) Glucose catabolism and TCA cycle (B) Methanol degradation (C) Respiratory chain components. (A) Dashed arrows denote reactions of the Entner Doudoroff (ED) pathway, PPP, Pentose Phosphate Pathway; DH, dehydrogenase, KDPG, 2-keto-3-deoxy-6-phosphocluconate. (B): H4MPTP, methylene tetrahydromethanopterin pathway; MtdA, methylene tetrahydromethanopterin dehydrogenase; FAE, formaldehyde activating enzyme; GSH-FAE, Glutathione dependent formaldehyde activating enzyme; FDH, formate dehydrogenase. (C) Q-pool, quinone pool. Gene numbers for all components or the different pathways are listed in the supplementary tables. Carbon metabolism—Table S2; Respiratory chain—Table S3; methanol metabolism—Table S4.
