Figure 1.
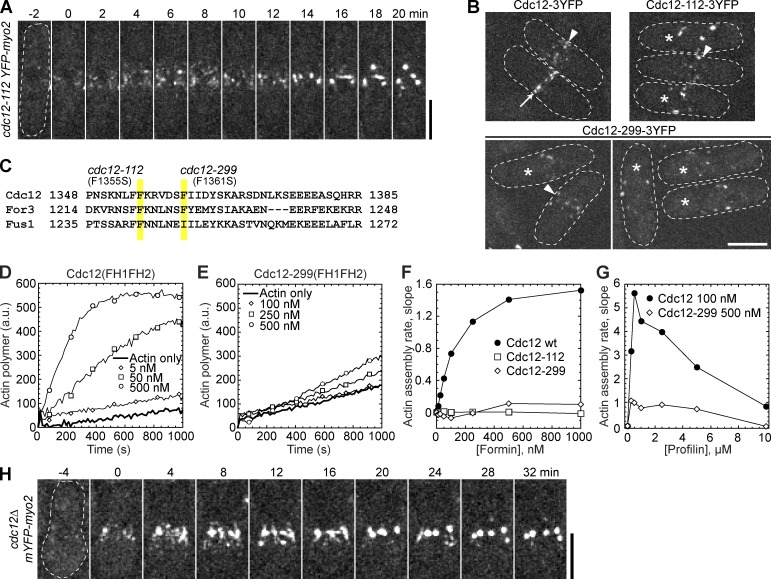
Condensation of cytokinesis nodes without functional formin Cdc12. (A and B) Cells were grown at 36°C for 2 h before imaging at 36°C. (A) Time course of Myo2 nodes condensing into clumps in cdc12-112 mutant. Time 0 is when nodes appear. (B) Localization of Cdc12, Cdc12-112, and Cdc12-299. Nodes (arrowheads), contractile rings (arrows), and cells with scattered clumps (asterisks) are marked. (C) Alignment of the last α-helix of the FH2 domains (αT; Xu et al., 2004) of S. pombe formins. Positions of point mutations in cdc12-112 and cdc12-299 are highlighted. (D–G) In vitro spontaneous assembly of 2.5 µM Mg-ATP actin monomers (10% pyrene labeled) using purified FH1-FH2 domains from wt and mutant Cdc12, in the absence (D–F) and presence (G) of profilin. The data are single representative experiments out of at least two repeats. (D and E) Time course of actin polymerization with the increasing concentrations of wt (D) or mutant (E) Cdc12. (F) Dependence of the actin assembly rate on formin concentration. (G) Dependence of Cdc12-mediated actin assembly rate on profilin concentration. (H) Time course of condensation of Myo2 nodes in cdc12Δ cells. Spores from cdc12Δ/cdc12+ diploid strain were germinated and grown at 25°C for 12 h before imaging at 25°C. Time 0 is when nodes appear. Bars, 5 µm.