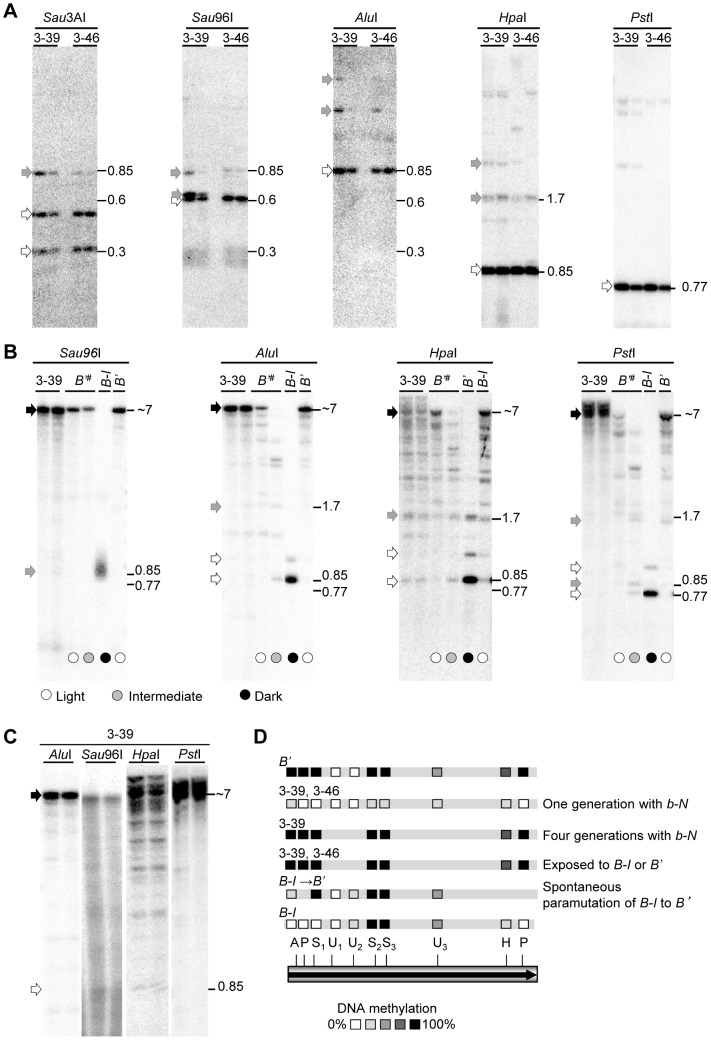
Figure 4. DNA methylation patterns in transgenic and endogenous b1 repeats.
Fragment sizes are indicated by numbers and are in kb. All DNA blots were hybridized with the full b1 repeat probe (Figure 3A). For all blots, genomic leaf DNA was cut with the methylation insensitive enzyme EcoRI to release the ∼7 kb fragment within which DNA methylation was assayed, and with the methylation sensitive enzymes indicated above each blot. Fragments resulting from complete digestion are indicated by open arrows, bands resulting from partial digestion (indicating partial DNA methylation) are indicated by gray arrowheads, while fragments that are the result of no digestion by methylation sensitive enzymes (indicating complete DNA methylation) are indicated by black arrowheads. Representative examples of DNA methylation patterns are shown. A. The progeny of two independent paramutagenic pBΔ transgenic events (3-39 and 3-46) were examined. The plants analyzed were the direct progeny of the primary transgenic plants and the transgenes were not yet exposed to B' or B-I. Results for representative b-N/b-N; TG/- plants (Figure 2A) are shown. B. B-I was exposed to the pBΔ transgenic event 3-39 for one generation, resulting in light pigmented plants, and then the transgene and the newly induced B' # were segregated away from each other. The transgene, B' # and control samples (B' and B-I) were assayed. Circles at the bottom of the lanes indicate plant pigment phenotypes. C. The pBΔ transgenic locus 3-39 was propagated for four generations in a neutral b-N background. D. Summary of the DNA methylation data for the transgenic 3-39 and 3-46 lines, plants in which B-I spontaneously paramutated to B' (Figure S4), and the previously determined B' and B-I patterns [21]. One and four generations with b-N indicates the 3-39 allele was propagated for one and four generations in the presence of neutral b-N alleles, respectively. The one repeat shown represents all seven repeats. Subscripts indicate specific recognition sites present more than once in each repeat. AluI (A), HpaI (H), PstI (P), Sau3AI (U) and Sau96I (S).