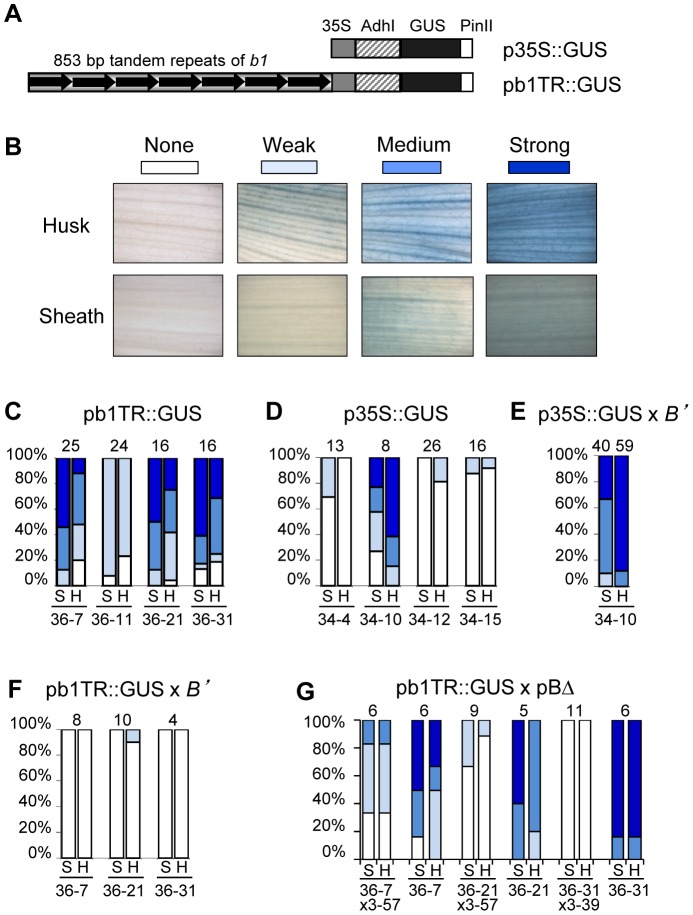
Figure 5. b1 tandem repeats are sufficient to mediate transcriptional activation in maize.
A. Drawing of the transgenic constructs used to assay transcriptional regulatory activity of the b1 repeats. The pb1TR::GUS construct carries the seven tandem b1 repeats (b1TR). The promoter is the minimal −90 bp Cauliflower Mosaic Virus 35S promoter (35S), which contains no enhancer sequences [28]. GUS is the beta-glucuronidase gene from E. coli [29]. PinII is the 3′ untranslated region from the potato proteinaseII gene [61]. The p35S::GUS construct, in which GUS expression is driven by the minimal −90 35S promoter, was used as a control. B. The scoring scale used to evaluate GUS expression levels in sheath and husk tissues of transgenic plants. Panels C–G show the percentage of transgenic plants with specific levels of GUS staining. The shades of blue correspond to the levels of GUS staining indicated in panel B; white signifies no staining. Sheath and husk tissues are denoted as S and H, respectively. The number of plants assayed is shown on the top while the transgenic events are shown below each pair of columns. Panels C and D show GUS staining levels for pb1TR::GUS and p35S::GUS in a neutral b1 background. Panels E and F show GUS staining levels for p35S::GUS and pb1TR::GUS transgene loci in the presence of the paramutagenic B' allele. Panel G shows GUS staining levels in two classes of progeny derived from crosses between GUS expressing pb1TR::GUS transgenic events and paramutagenic pBΔ events. One class carries both a pb1TR::GUS and pBΔ transgene, the second class carries only a pb1TR::GUS transgene. All transgenic loci shown in panels C–G were in a hemizygous state.