Figure 6. Northern blot analysis of repeat siRNAs in transgenic plants.
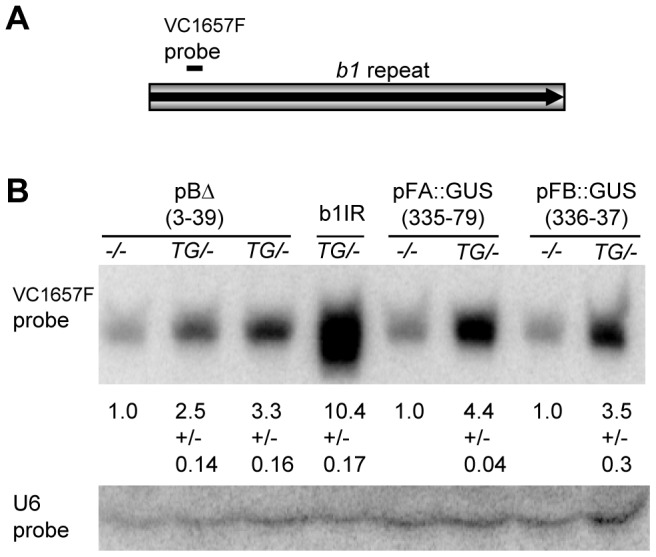
A. The levels of the b1 tandem repeat siRNAs were detected with the indicated oligo probe (VC1657), which hybridizes to the FA part of the repeat. B. Genotypes of the plants used for the analysis are shown above the blot with −/− denoting the absence of the transgene and TG/- indicating the presence of one copy of the transgene locus. b1IR stands for 35S::b1IR. The RNA levels were detected by hybridization with 32P labelled DNA/LNA oligonucleotide probes as described by [24]. The levels of the b1 tandem repeat siRNAs detected with the VC1657 probe were normalized to U6 RNA levels, which served as a loading control [24]. The average abundances of b1 repeat siRNAs are presented relative to the levels in their non-transgenic sibling plants, which were set to 1.0; +/− indicates the standard deviation. A 35S::b1IR transgene that produces high levels of siRNAs was used as a positive control for hybridization and is described in [24]. Transgenes pBΔ 3-39, pFA::GUS and pFB::GUS are described in Figure 1 and Figure S6, respectively.
