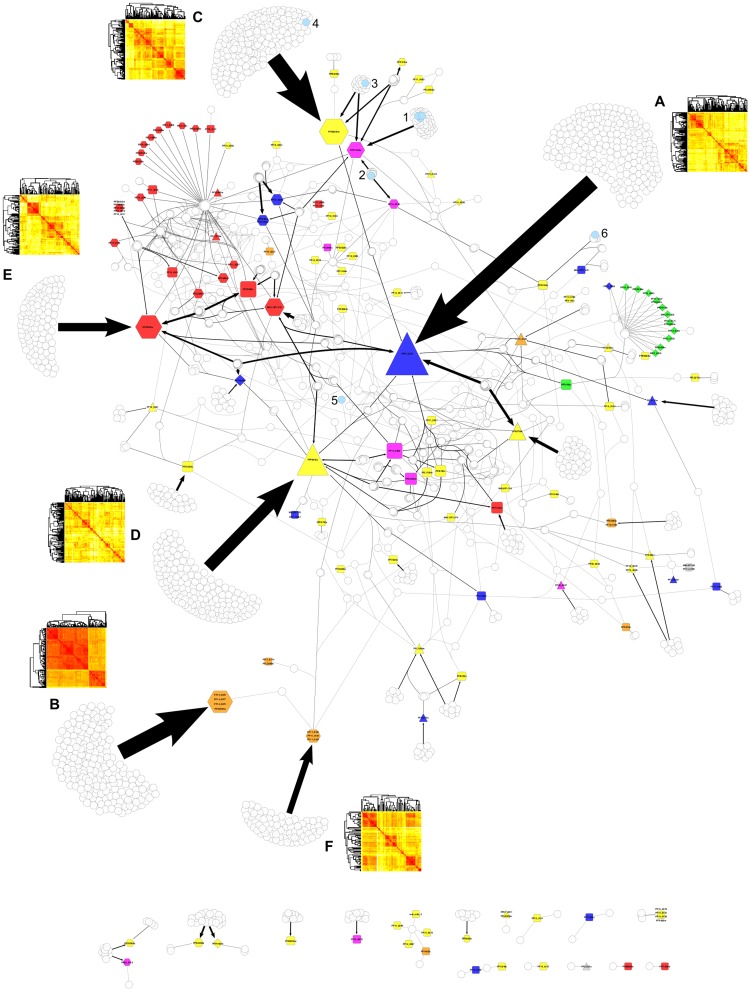
Figure 4. Drug-target network of 1,908 active compounds predicted for 147 P. falciparum proteins.
Node colors encode target families (red: kinase; orange: protease; yellow: other enzyme; green: ribosomal protein; magenta: transporter/channel; blue: other protein; grey: unknown function), node shapes encode prioritization (hexagon: high priority; triangle: high affinity target; diamond: enriched target; square: other predictions). Targets from the same orthologous group are merged into one common node. Compounds are shown as white circles and grouped according to their target profiles. Edge width corresponds to the number of compounds in the respective group. Clinically used drugs are highlighted in light blue. The numbering corresponds to chloroquine, mefloquine, and artemether (1), quinine (2), pyrimethamine (3), proguanil (4), sulfadoxin (5), and atovaquone (6). Capital letters are used to identify the target hubs of Hsp90 (A), plasmepsin I, II, IV, and VI (B), bifunctional dihydrofolate reductase-thymidylate synthase (C), acyl-CoA synthetase (D), serine/threonine protein kinase ARK2 (E), and falcipain 2a, 2b, and 3 (F).