Table 1. Experimental validation of the binding events predicted by dPeak analysis of 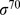 PET ChIP-Seq data.
PET ChIP-Seq data.
| Genea | Predicted position | True positionb | Distance | Primerb | Conditionc |
| yejG | 2,276,288 | 2,276,299 | 11 | P 1 | Aerobic |
| yejG | 2,276,432 | 2,276,419 | 13 | P 2 | Aerobic |
| spr | 2,267,945 | 2,267,942 | 3 | P 1 | Aerobic |
| spr | 2,267,825 | 2,267,833 | 8 | P 2 | Aerobic |
| dcuA | 4,364,876 | 4,364,866 | 10 | P 1 | Anaerobic |
| dcuA | 4,364,975 | 4,364,974 | 1 | P 2 | Anaerobic |
| aroL | 405,583 | 405,579 | 4 | P 1 | Anaerobic |
| aroL | 405,489 | 405,504 | 15 | P 2 | Anaerobic |
| serC | 956,823 | 956,802 | 21 | P 1 | Aerobic |
| serC | 956,789 | (Not validated) | N/A | Aerobic | |
| hybO | 3,144,382 | 3,144,385 | 3 | P 1 | Anaerobic |
| hybO | 3,144,438 | (Not validated) | N/A | Anaerobic | |
| ybgI | 742,036 | 742,030 | 6 | P 1 | Aerobic |
| ybgI | 741,859 | 741,874d | 15 | P 1 | Aerobic |
| ptsG | 1,157,005 | 1,156,989 | 16 | P 1 | Aerobic |
| ptsG | 1,156,866 | 1,156,849d | 17 | P 1 | Aerobic |
The genes with promoters harboring the predicted binding events.
The true positions were determined by primer extension experiments (Figure 5A).
The conditions under which binding events are validated.
We report results based on the RegulonDB annotations for ybgI and ptsG genes as the primer extension products for these genes were too large to accurately map with the sequencing ladder.
