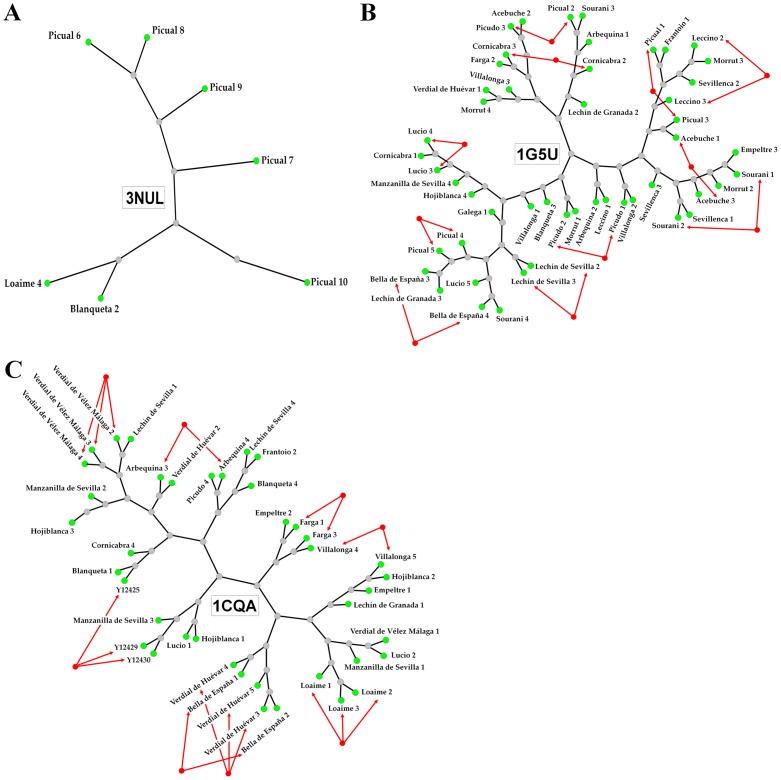
Figure 4. Profilin conservational analysis.
Consurf-conservational analysis of profilin proteins showed in three individual views rotated 90° for the PDB models A) 1g5uA, B) 1cqa, and C) 3nul, respectively. The conserved and variable residues are presented as space-filled models and colored according to the conservation scores. The strictly conserved and variable residues are depicted in purple and blue, respectively. Red dotted circles and red arrows point a detailed of the plant specific solvent-filled. The sequence of the protein is depicted with the evolutionary rates color-coded onto each site. The residues of the query sequence are always numbered starting from 1. The predicted burial status of the site (i.e. “b”-buried vs. “e”-exposed) is annotated under the sequence. Residues predicted to be structurally and functionally important, “s” and “f”, are also pointed out under the sequence. Amino-acid sites categorized as “Insufficient data” are colored in yellow, indicating that the calculation for these sites were generated using only a few of the homologous sequences. Orange, light blue and purple starts highlight the key amino acids implicated in the interaction with actin, PLP and PIP, respectively. Red lines under the sequences represent the profilin characteristic motif, which define this family of proteins. C = conserved, V = variable, U = undefined.