FIGURE 2.
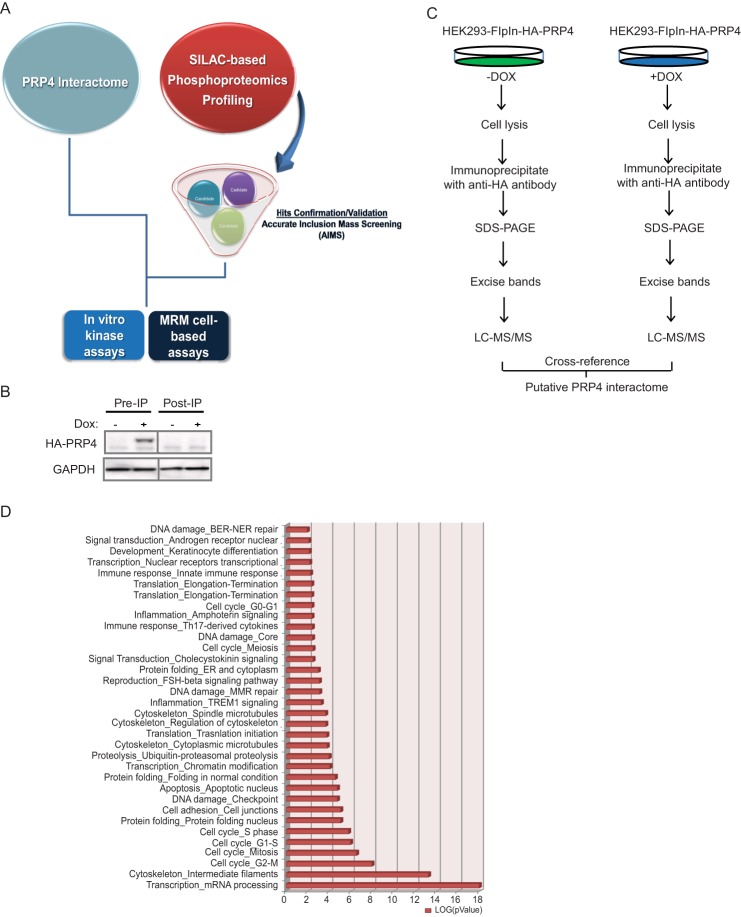
The PRP4 interactome. A, PRP4 substrate hunting strategy. PRP4 interactome was characterized by LC-MS/MS with the assumption that PRP4 substrates should be physically associated with PRP4. In parallel, a SILAC-based phosphoproteomics profiling experiment coupled with inducible knockdown of PRP4 was performed, and results were validated by AIMS. The interactome data were cross-referenced with the AIMS data to narrow down the candidate list. MRM-based mass spectrometry and PRP4 in vitro kinase assays were utilized to further validate the selected PRP4 substrate. B, successful depletion of exogenously expressed HA-tagged PRP4 by immunoprecipitation (IP) with anti-HA antibody. C, immunoprecipitation-MS workflow for characterization of PRP4 interactome. Exogenous expression of HA-tagged PRP4 kinase was induced by addition of doxycycline (DOX) and immunoprecipitated by the anti-HA antibody after lysis. The PRP4-associated proteins were separated by SDS-PAGE followed by in-gel digestion and mass spectrometry identification. For control, an identical experiment was performed using lysates without the induced expression of HA-PRP4. D, functional classification of PRP4 binding partners using GeneGo software based on universal gene ontology annotation terms. The top six functional categories are enriched with proteins associated with mRNA processing and cell cycle regulation. All categories presented in the graph have p values ≤0.01. BER-NER, base excision repair-nucleotide excision repair; ER, endoplasmic reticulum; MMR, mismatch repair.