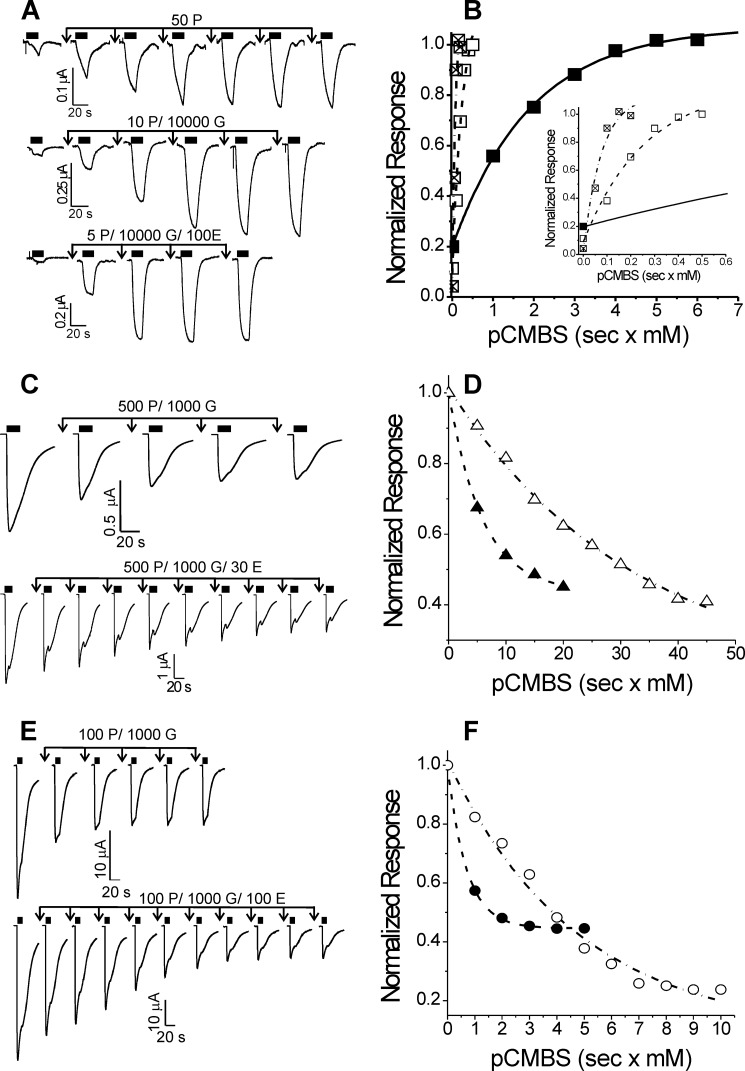
FIGURE 6.
Etomidate effects on cysteine modification in GABAA receptors with α1Q229C, α1L232C, and α1T237C mutations. Left-hand panels display current traces from oocytes expressing mutant GABAA receptors. Currents were activated with GABA, indicated by solid black bars above each trace. Downward arrows indicate exposures to pCMBS alone or with GABA or with GABA plus etomidate, followed by washout. Modification conditions (P, pCMBS; G, GABA; E, etomidate) for each set of traces are indicated in micromolar above each set of arrows. Right-hand panels show normalized peak current data from the traces shown on the left plotted against cumulative pCMBS exposure (s × mm). Lines through plotted symbols represent least squares fits to single exponentials. A, α1Q229Cβ2γ2L channels. Currents were stimulated with 50 μm GABA (EC10). B, solid squares, modification with pCMBS alone (fitted rate = 520 m−1 s−1); open squares, modification with pCMBS plus GABA (fitted rate = 3280 m−1 s−1); crossed squares, modification with pCMBS plus GABA and etomidate (fitted rate = 6500 m−1 s−1). Inset, rate analyses are shown with a magnified time base. C, α1L232Cβ2γ2L channels. Currents were stimulated with 1 mm GABA (EC100). D, solid triangles, modification with pCMBS plus GABA (fitted rate = 170 m−1 s−1); open triangles, modification with pCMBS plus GABA and etomidate (fitted rate = 34 m−1 s−1). E, α1T237β2γ2L channels. Currents were stimulated with 1 mm GABA (EC100). F, solid circles, modification with pCMBS plus GABA (fitted rate = 1450 m−1 s−1); open circles, modification with pCMBS plus GABA and etomidate (fitted rate = 210 m−1 s−1).