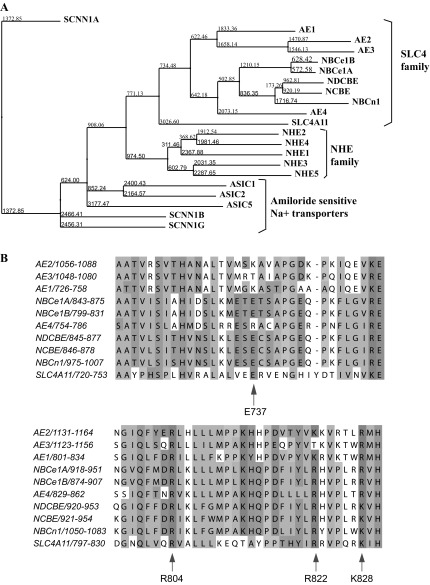
Fig. 14.
Sequence analysis of SLC4A11. A: phylogenetic tree showing relationship between SLC4, NHE, and Amiloride-sensitive Na+ transporter families of ion transporters. Sequences were aligned with ClustalW2 program and the alignment was loaded in Jalview 2.7 software (http://jalview.org) to generate the phylogenetic tree showing evolutionary relationship between the proteins. Numbers at the nodes and the lengths of the lines indicate relative evolutionary distance. B: SLC4 family alignment of the first hinge loop (top) and the second hinge loop (bottom). This 1st hinge loop is located in the EL5 of SLC4A11 between TM9 and TM10. The 1st hinge loop is postulated to have sodium attracting amino acids. E737 (Glutamine, net negative charge) is conserved in all members of the SLC4 family that transport sodium and also in SLC4A11. The 2nd hinge loop is located in the TM12 domain of SLC4A11. This region it is postulated to have anion attracting motifs. R804, R822, and K828 (arginine and lysine, net positive charge) are conserved in SLC4A11.