Abstract
Background:
The TP53 pathway is frequently inactivated in human cancers. PICT1 (also known as GLTSCR2) is a novel regulator of the MDM2-TP53 pathway via its interaction with the ribosomal protein RPL11 in the nucleolus. However, the clinical significance of PICT1 in gastric cancer remains unknown.
Methods:
To evaluate PICT1 function, we used shRNA to inhibit PICT1 expression in gastric cancer cells that expressed wild-type TP53. PICT1 expression and TP53 mutation status were quantified in 110 cases of primary gastric cancer to explore the impact of PICT1 expression levels on gastric cancer.
Results:
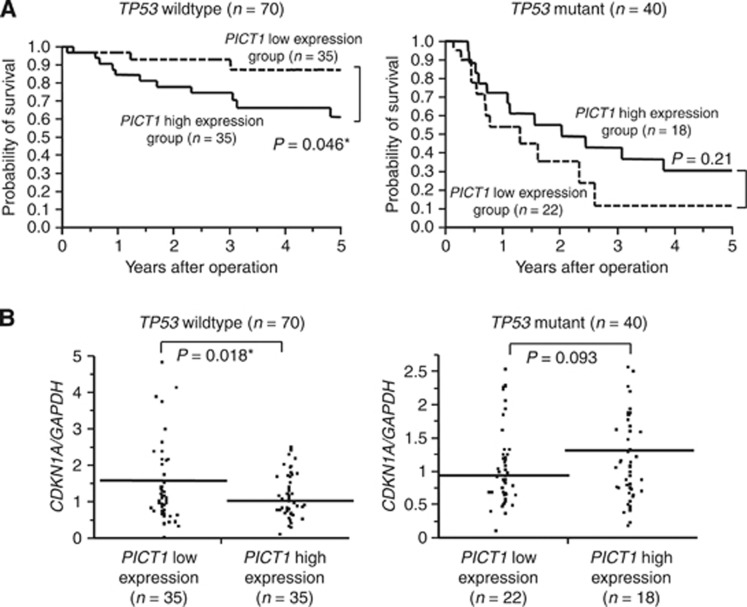
Deficiency of PICT1 significantly impaired cell proliferation and colony formation via TP53-mediated cell cycle arrest. Following induction of PICT1 deficiency, RPL11 translocated out of the nucleolus. Of the 110 gastric cancer samples tested, 70 (63.6%) and 40 (36.4%) tumours expressed wild-type and mutant TP53, respectively. In gastric cancer patients with wild-type TP53 tumours, patients with relatively low PICT1 expression levels had a better prognosis compared with high expression level patients (P=0.046).
Conclusion:
The findings suggest that PICT1 has a crucial role in gastric cancer progression by regulating the MDM2-TP53 pathway through RPL11. Clinically, PICT1 expression is a novel prognostic parameter in gastric cancer patients with wild-type TP53 tumours.
Keywords: PICT1, TP53, gastric cancer, RPL11, MDM2, GLTSCR2
Gastric cancer is one of the most common malignant tumours in Japan, and it is strongly associated with Helicobacter pylori infection. H. pylori infection inhibits TP53 function by the activation of MDM2, which promotes TP53 degradation (Wei et al, 2010). TP53 is a major tumour suppressor and transcription factor that induces cell cycle arrest, apoptosis and senescence. More than 30% of gastric cancer patients harbour TP53 mutations, and TP53 inactivation has an important role in tumorigenesis and tumour progression (Fenoglio-Preiser et al, 2003). The expression of TP53 is induced by DNA damage, and TP53 is degraded by MDM2 – an E3 ubiquitin (Ub) ligase (Haupt et al, 1997; Kubbutat et al, 1997). Therefore, the activation of MDM2 leads to a reduction in TP53 levels and results in tumour progression (Toledo and Wahl, 2006). The MDM2-TP53 pathway is also regulated by certain ribosomal proteins such as RPL5, RPL11, RPL23, and RPS7, which, in response to the nucleolar stress, can bind to MDM2 in the nucleoplasm and inhibit its function (Zhu et al, 2009).
PICT1 (also known as GLTSCR2) was considered to be a tumour suppressor because of its localisation at 19q13 (a region that is frequently deleted in human malignant tumours) and its ability to stabilise PTEN (Okahara et al, 2004; Okahara et al, 2006; Yim et al, 2007). However, using tetracycline-induced Pict1-deficient ES cells, we recently showed that Pict1 bound to ribosomal protein Rpl11 in the nucleolus. Loss of Pict1 led to the release of Rpl11 from the nucleolus, whereupon it translocated to the nucleoplasm to inhibit Mdm2, which resulted in stabilisation and accumulation of the p53 protein (Sasaki et al, 2011). Moreover, in colorectal cancer (CRC) and esophageal squamous cell carcinoma (ESCC) patients with wild-type TP53 tumours, patients in the PICT1 low-expression group had a significantly better prognosis. Thus, PICT1 appears to act as an oncogene in cells by inactivation of the TP53 pathway. However, little is known about regulation of PICT1 of the MDM2-TP53 pathway in gastric cancer cells and its clinical significance in human gastric cancer cases.
Therefore, we examined the controversial ability of PICT1 to regulate the MDM2-TP53 pathway, especially in gastric cancer cells. In addition, we characterised the clinical significance of PICT1 expression levels in 110 gastric cancer cases, establishing PICT1 status as a bona fide prognostic marker in gastric cancer.
Materials and methods
Gastric cancer cell in vitro analysis
AGS cells were obtained from the American Type Culture Collection (ATCC). These cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Gibco-BRL, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (FBS), 100 units ml−1 penicillin, and 100 μg ml−1 streptomycin in a humidified incubator at 37 °C under 5% CO2.
Quantitative real-time PCR
PICT1, TP53, MDM2, and CDKN1A mRNA levels were quantified using LightCycler 480 Probes Master kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's protocol with specific primers shown in the Supplementary Table 1. Gene-expression levels were normalised with respect to GAPDH.
Immunoblotting analysis of TP53, CDKN1A, BAX, and PICT1 protein expression
We carried out immunoblotting using a standard protocol and primary antibodies against PICT1, TP53 (Dako, A/S, Glostrup, Denmark), CDKN1A, and BAX (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The proteins were detected using HRP-conjugated secondary antibodies (GE Healthcare Bioscience, Tokyo, Japan). The PICT1 antibody was affinity-purified from antisera as previously described (Okahara et al, 2005). β-actin levels (Santa Cruz Biotechnology) were used as a loading control.
Ubiquitin ligase activity assays
We assayed Ub ligase activity as described previously (Furukawa et al, 2000; Feng et al, 2005) with minor modifications. AGS cells in 9-cm diameter dishes were infected for 3 days with lentiviral PICT1-shRNA as described (Inoue-Narita et al, 2008). Infected cells were cotransfected with pCAG-HA-Ub (3.5 μg), pcDNA3.1-Myc-TP53 (3.5 μg), and using FuGENE HD (Roche Applied Science). The total amount of plasmid DNA in each transfection was adjusted to 10.5 μg using empty pcDNA3.1 vector as needed. Two days post transfection, cells were treated with 20 μM proteasome inhibitor MG132 (Sigma, St Louis, MO, USA) for 6 h. Cells were boiled for 10 min in 100 μl SDS lysis buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1% SDS, and 1 mM dithiothreitol), and lysates were diluted in 1 ml 0.5% Nonidet P40 buffer. Ubiquitinated protein was immunoprecipitated (IP) from cell lysates using 2 μg HA-specific antibody (Ab) (Roche Applied Science), resolved by SDS-PAGE, and immunoblotted (IB) with ubiquitinated TP53-specific Ab (Santa Cruz Biotechnology).
Cellular proliferation assays
AGS cells were seeded on 96-well plates at a density of 1 × 103 cells per well. After 24 h, cells were infected with lentiviral PICT1-shRNA. Cell proliferation was evaluated with the MTT assay using Cell Proliferation Kit I (Roche Applied Science) following the manufacturer's protocol.
Tumour transformation assay
Forty-eight hours after lentiviral PICT1-shRNA infection, AGS cells were trypsinised and well mixed with prewarmed RPMI1640 medium containing 10% FBS and 0.33% agar (Difco Noble Agar, BD Biosciences, San Jose, CA, USA). Two milliliter of this mixture from each group was seeded into a 60-mm plate precoated with 3 ml RPMI1640 medium containing 0.5% agar. The plates were incubated at 37 °C and 5% CO2 for 14 days. The number of colonies was counted under an inverted microscope.
Cell cycle assays
Seventy-two hours after lentiviral PICT1-shRNA infection, AGS cells were fixed in 70% ethanol at −20 °C and resuspended in PBS containing five μg ml−1 propidium iodide (PI) and 0.25 mg ml−1 RNase A. Data were collected on a FACSVantage flow cytometer (BD Biosciences) and analysed with FlowJo v9.3.3 (TreeStar, Ashland, OR, USA).
Confocal microscopy
Lentiviral PICT1-shRNA and RPL11-DsRed expression plasmids were cotransfected into AGS cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Seventy-two hours after lentiviral PICT1-shRNA infection, cells were fixed in 4% formaldehyde and dehydrated with cold methanol-acetone (1 : 1). Fixed cells were immunostained with anti-NPM antibody (Sigma), followed by incubation with Alexa350-labelled secondary antibodies (Invitrogen). Cells were then examined using confocal microscopy (LSM5, Carl Zeiss, Jena, Germany).
Apoptosis assays with anticancer drugs
Forty-eight hours after lentiviral PICT1-shRNA infection, AGS cells were treated with camptothecin and cisplatin (both at 10 μg ml−1) for 24 h and were stained with annexin V-APC (BD Biosciences). Annexin-V-positive cells were measured using a FACSVantage flow cytometer (BD Biosciences) and analysed using FlowJo v 9.3.3 (TreeStar).
Gene-set-enrichment analysis of gastric cancer cell lines with PICT1 expression
Gene-expression profiles and TP53 status of 34 gastric cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al, 2012). Of the 34 gastric cancer cell lines, 11 (32.4%) and 23 (67.6%) tumours expressed wild-type and mutant TP53, respectively. We used gene-set-enrichment analysis (GSEA) to clarify the correlation between PICT1 expression and gene signatures for each TP53 status group (Subramanian et al, 2005). A GSEA algorithm was applied to identify the enrichment of specific functions in the list of genes preranked according to their P-value for the test of differences in expression between cell lines. The statistical significance of the enrichment score was calculated by permuting the genes 1000 times as implemented in the GSEA software. To collapse each probe set on the array to a single gene, the probe with the highest variance among multiple probes that corresponded to the same gene was selected. Gene sets were derived from the REACTOME pathway database (Croft et al, 2011).
Patients and samples
One hundred ten gastric cancer samples were obtained during surgery and used after obtaining informed consent. All patients underwent resection of the primary tumour at the Kyushu University Beppu Hospital between 1992 and 2000. None of the patients received neoadjuvant chemotherapy or radiotherapy before surgery. Resected cancerous tissues were immediately cut and stored in RNA later (Applied Biosystems, Foster City, CA, USA), frozen in liquid nitrogen, and kept at −80 °C for subsequent RNA and DNA extraction. Following isolation of RNA, cDNA was synthesised from 8.0 μg total RNA as described previously (Yokobori et al, 2009). Genomic DNA was extracted from cancer tissues using the QIAamp DNA mini kit according to the manufacturer's protocol (QIAGEN, Valencia, CA, USA), followed by direct DNA sequencing.
DNA sequencing of TP53
Genomic DNA and RNA were extracted from the 110 gastric cancer tissues and their TP53 status determined using direct DNA sequencing of TP53 exons five to eight, the area where most TP53 mutations occur. Exons five to eight of the TP53 gene were amplified and sequenced using BigDye Terminator v3.1 (Applied Biosystems) as previously described (Yokobori et al, 2009).
Statistical analyses
Differences between two groups were estimated with Student's t-test and χ2 test. Overall survival curves were plotted according to the Kaplan–Meier method, with the log-rank test applied for comparison. Survival was measured from the day of the surgery. All differences were statistically significant at the level of P<0.05. Statistical analyses were performed using the JMP 5 for Windows software package (SAS Institute).
Results
PICT1 deficiency resulted in TP53 protein accumulation and inhibited proliferation of gastric cancer cells expressing wild-type TP53
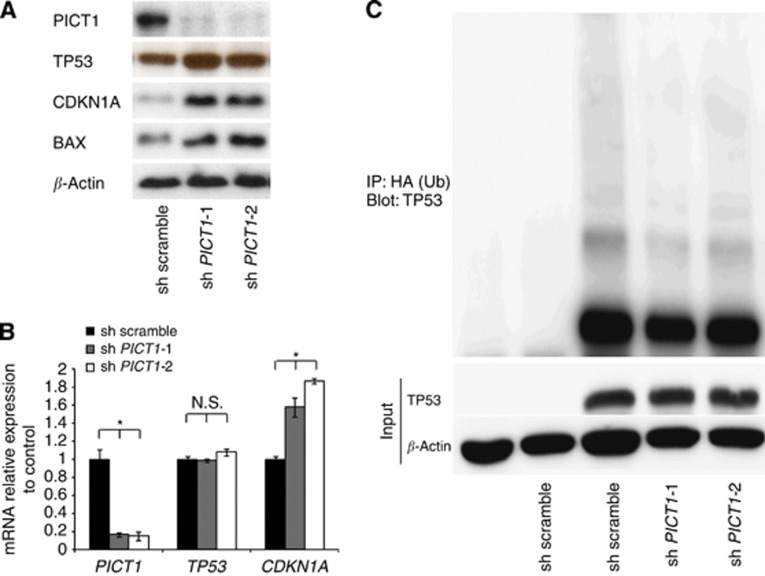
We performed a detailed analysis of the function of PICT1 in the gastric cancer cell line AGS that expresses wild-type TP53. Following treatment with lentivirus PICT1-shRNA, a marked reduction in PICT1 protein was confirmed (Figure 1A). PICT1 deficiency in AGS cells induced TP53 protein accumulation and upregulation of CDKN1A and BAX, which are major TP53 transcription targets (Figure 1A) (el-Deiry et al, 1993; Miyashita and Reed, 1995). The accumulation of TP53 was not observed in two gastric cancer cell lines, NUGC3 and MKN7, which carry mutant TP53 (Supplementary Figure 1A). The data demonstrated that there was no significant change in TP53 mRNA levels or an increase in CDKN1A mRNA levels (Supplementary Figure 1B). In ES cells, Pict1 deficiency prevented p53 degradation by decreasing Mdm2-mediated ubiquitination (Sasaki et al, 2011). Thus, we next asked whether reducing PICT1 expression by cancer cells affected TP53 degradation by ubiquitination. We transfected AGS cells with plasmids expressing hemagglutinin-tagged Ub (HA-Ub) and TP53, and we carried out immunoprecipitation and immunoblotting to detect ubiquitinated TP53 (Ub-TP53). As anticipated, when we infected TP53-expressing AGS cells with a lentivirus expressing PICT1-shRNA, the laddering was less intense (Figure 1C).
Figure 1.
PICT1 deficiency resulted in TP53 protein accumulation and prevented TP53 ubiquitination. (A) Western blot analysis of PICT1, TP53, CDKN1A, and BAX in AGS cells infected with lentivirus expressing scrambled shRNA or PICT1-shRNA-1 or -2. β-actin levels were used as a loading control. (B) Quantitative real-time PCR of PICT1, TP53, and CDKN1A in AGS cells infected with lentiviral PICT1-shRNA. Gene-expression levels were normalised with respect to GAPDH. Error bar: mean±s.d. *P<0.05, significant difference from controls. (C) Immunoblot of AGS cells transfected with the indicated plasmids and treated with MG132 (20 μM). Lysates were immunoprecipitated and immunoblotted with antibodies to HA (ubiquitin, Ub) and ubiquitinated TP53.
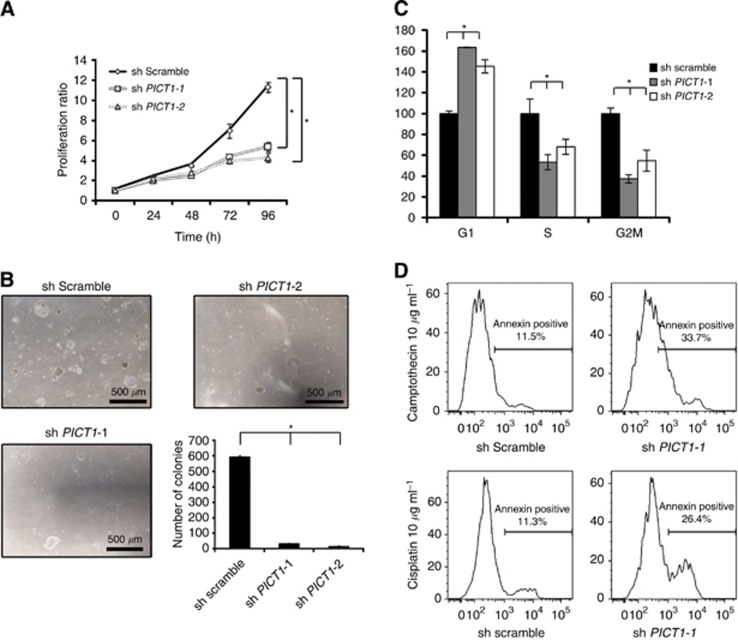
Next, to analyse the phenotype induced by PICT1 deficiency, we performed a cell-proliferation assay. PICT1 deficiency inhibited anchorage-dependent growth (Figures 2A and B) and also decreased the proportion of cells in the S and G2/M phases, as measured by a cell cycle assay, which indicated that PICT1 depletion induced G1 arrest (Figure 2C) in AGS. In contrast, PICT1 deficiency in NUGC3 and MKN7, which carry mutant TP53, did not inhibit cell growth (Supplementary Figure 1B). Moreover, PICT1 deficiency increased the number of apoptotic cells after treatment with antigastric cancer drugs, such as cisplatin and camptothecin (Figure 2D). Thus, inhibition of PICT1 expression induced TP53 accumulation and inhibited transformation in not only ES cells but also human gastric cancer cells.
Figure 2.
PICT1 deficiency inhibited proliferation of gastric cancer cells expressing wild-type TP53. (A) Cell proliferation was evaluated using MTT assay. Error bar: mean±s.d. *P<0.05, significant difference from controls. (B) Tumour transformation assay. The number of colonies were counted under an inverted microscope. Representative images of AGS colonies 14 days post transfection with shRNA-targeting PICT1 or scrambled (control) are shown. Graph shows the number of colonies in each sample. Error bar: mean±s.d. *P<0.05, significant difference from controls. (C) Cell cycle assay. Graph shows flow cytometric profiles of the percentage of cells in each cell cycle phase. Error bar: mean±s.d. *P<0.05, significant difference from controls. (A–C) Results represent three trials. (D) PICT1 deficiency increased apoptosis in gastric cancer cells treated with anticancer drugs in gastric cancer cells. Forty-eight hours after lentiviral PICT1-shRNA infection, AGS cells were treated with camptothecin and cisplatin (each 10 μg ml−1) for 12 h and were stained with annexin V-APC (BD Biosciences).
PICT1 deficiency induced translocation of RPL11 out of the nucleolus
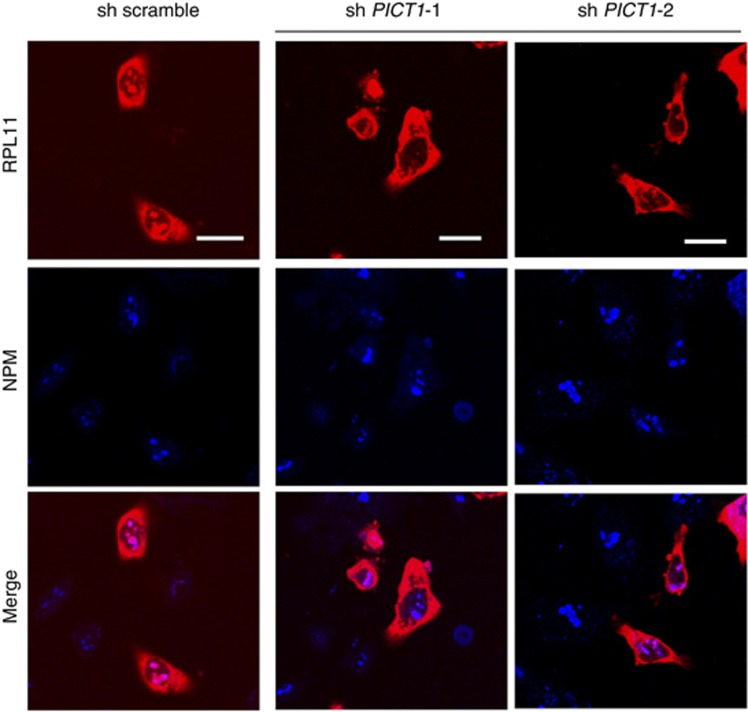
In ES cells, Pict1 binds to Rpl11 in the nucleolus, whereas in the absence of Pict1, Rpl11 is released from the nucleolus to the nucleoplasm (Sasaki et al, 2011). The released Rpl11 binds to Mdm2, which inhibits Mdm2-mediated ubiquitination of p53. Translocation of Rpl11 out of the nucleolus is one of the most important events leading to p53 accumulation in Pict1-deficient ES cells (Sasaki et al, 2011). Thus, we next explored whether PICT1 depletion in cancer cells affected RPL11 localisation by cotransfecting gastric cancer cells with lentiviral PICT1-shRNA and RPL11-DsRed expression vectors. RPL11 appeared in both the nucleolus and the cytoplasm in cells transfected with scrambled shRNA but translocated out of the nucleolus in cells transfected with PICT1-shRNA (Figure 3). Thus, PICT1 appears to be essential for the nucleolar localisation of RPL11 in gastric cancer cells.
Figure 3.
PICT1 deficiency induced translocation of RPL11 out of the nucleolus. Confocal microscopy of AGS cells cotransfected with lentiviral PICT1-shRNA and RPL11-DsRed expression plasmid. Scale bars, 5 μm. Confocal microscopy of AGS cells cotransfected with lentiviral PICT1-shRNA and RPL11-DsRed expression plasmids. Endogenous nucleophosmin (NPM) was detected using antibody to NPM (blue). Cell fluorescence at 72 h is shown. Scale bars, 5 μm.
GSEA revealed correlations between PICT1 and TP53 pathways in TP53 wild-type gastric cancer cell lines
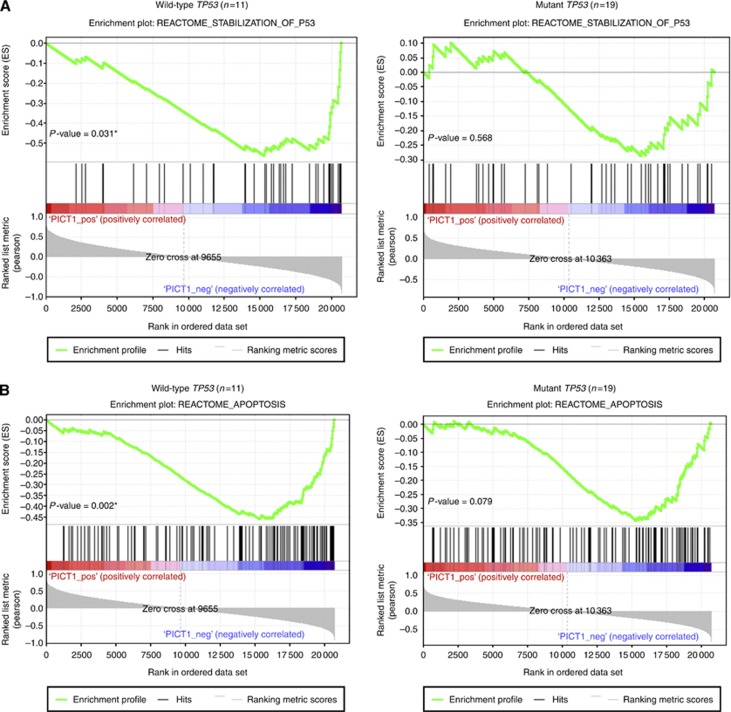
We used the array data of gastric cancer cell lines from CCLE and asked whether PICT1 expression was highly correlated with previously curated gene-expression signatures (Croft et al, 2011). In these cell lines, 11 lines (including AGS) have wild-type TP53 and 19 cell lines have mutated TP53 (Barretina et al, 2012). We used GSEA to examine possible correlations between PICT1 expression and TP53-associated pathways for each TP53 status groups. Although the gene signature, REACTOME_STABILIZATION_TP53, was not correlated with PICT1 expression in TP53-mutated cell lines (P=0.568), it was significantly negatively correlated with PICT1 expression in TP53 wild-type cell lines (P=0.031) (Figure 4A). In addition, the gene pathway, REACTOME_APOPTOSIS, was negatively correlated with PICT1 expression only in TP53 wild-type gastric cancer cell lines (P=0.002) (Figure 4B). These results suggested that PICT1 induces TP53 ubiquitination and apoptosis not only in AGS cell lines but also in other TP53 wild-type gastric cancer cell lines.
Figure 4.
GSEA in gastric cancer cell lines revealed correlations between PICT1 expression and TP53 stabilisation. Enrichment plots of expression signatures of REACTOME_STABILIZATION_OF_P53 and REACTOME_APOPTOSIS. The barcode plot indicates the position of the genes in each gene set; red and blue colours represent positive and negative Pearson's correlation with PICT1 expression, respectively. These gene-expression signatures were obtained from the REACTOME database (Croft et al, 2011). (A) GSEA for the gene signature of the REACTOME_STABILIZATION_OF_P53. (B) GSEA for the signature of REACTOME_APOPTOSIS. *P<0.05, significant difference was predicted by statistic analysis.
Low PICT1 expression levels were associated with a better prognosis in gastric cancer patients with wild-type TP53 tumours
We investigated whether PICT1 also behaved as an oncogene in clinical samples. Genomic DNA and RNA were extracted from 110 gastric cancer tissues and their TP53 status was determined by direct DNA sequencing of TP53 exons five to eight. PICT1 expression levels in these samples were determined using qRT–PCR. Of the 110 gastric cancer samples tested, 70 (63.6%) and 40 (36.4%) tumours had wild-type and mutant TP53, respectively. We divided the two groups according to their PICT1 expression level (PICT1 high-expression group, PICT1/GAPDH>1; low-expression group, PICT1/GAPDH<1, Supplementary Figure 2A). In gastric cancer cases with wild-type TP53 tumours (n=70), the PICT1 high-expression group (n=35) had a poorer prognosis for overall survival as compared with the low-expression group (n=35, P=0.046) (Figure 5A, left). However, for mutant TP53 (n=40) and total gastric cancer cases (n=110), PICT1 expression levels did not correlate with overall survival (Figure 5A, right, Supplementary Figure 2B). Next, we investigated whether MDM2 was a prognostic marker in gastric cancer cases. There was no significant difference between the MDM2 high-expression group and the low-expression group in either wild-type TP53 or mutant TP53 gastric cancer cases (Supplementary Figure3).
Figure 5.
Low PICT1 expression levels were associated with a better prognosis in gastric cancer patients with wild-type TP53 tumours. (A) Kaplan–Meier overall survival curves for 110 gastric cancer patients according to PICT1 level. Left, patients with wild-type TP53 tumours (n=70); Right, patients with mutant TP53 tumours (n=40). A log-rank test was performed to determine significance. (B) Quantitative real-time PCR analysis of CDKN1A in 110 gastric cancer tissues and classification based on PICT1 level (PICT1/GAPDH=1). Horizontal line: borderline of PICT1 high or low. Left, wild-type TP53 tumours (n=70); Right, mutant TP53 tumours (n=40). A Student's t-test was performed to determine significance. *P<0.05, significant difference was predicted by statistic analysis.
Clinicopathologic variables of PICT1 expression in gastric cancer cases
We also analysed the association between PICT1 expression levels and clinicopathologic factors in gastric cancer patients with wild-type TP53 tumours and found that PICT1 expression was significantly associated with tumour depth (P=0.03, Table 1). Moreover, to clarify the relationship between PICT1 expression and the function of TP53 in gastric cancer cases, we examined the association between PICT1 and CDKN1A expression levels using qRT–PCR. In wild-type TP53 cases, the PICT1-low-expression group (n=35) showed significantly higher CDKN1A expression than the high-expression group (n=35) (P=0.018, Figure 5B, left). In contrast, for TP53-mutant cases, there were no significant differences between PICT1 and CDKN1A expression levels (Figure 5B, right).
Table 1. PICT1 expression and clinocopathologic factors in gastric cancer patients with wild-type TP53.
| |
PICT1
low expression (n=35) |
PICT1
high expression (n=35) |
|
||
|---|---|---|---|---|---|
| Factors | Number | % | Number | % | P-value |
| Age (mean±s.d.) |
66.5±2.00 |
|
64.5±2.00 |
|
0.477 |
|
Gender | |||||
| Male | 22 | 62.9 | 21 | 60.0 | 0.806 |
| Female |
13 |
37.1 |
14 |
40.0 |
|
|
Histological grade | |||||
| Well and moderately | 17 | 48.6 | 18 | 51.4 | 0.811 |
| Poorly and Signet |
18 |
51.4 |
17 |
48.6 |
|
|
Tumour deptha | |||||
| m, sm, mp | 21 | 60.0 | 12 | 34.3 | 0.03* |
| ss, se, si |
14 |
40.0 |
22 |
65.7 |
|
|
Lymph node metastasis | |||||
| Absent | 16 | 45.7 | 17 | 48.6 | 0.811 |
| Present |
19 |
54.3 |
18 |
51.4 |
|
|
Lymphatic invasion | |||||
| Absent | 13 | 37.1 | 9 | 25.7 | 0.302 |
| Present |
22 |
62.9 |
26 |
74.3 |
|
|
Venous invasion | |||||
| Absent | 32 | 91.4 | 26 | 74.3 | 0.052 |
| Present |
3 |
8.6 |
9 |
25.7 |
|
|
Peritoneal dissemination | |||||
| Absent | 34 | 97.1 | 30 | 85.7 | 0.075 |
| Present |
1 |
2.9 |
5 |
14.3 |
|
|
Stage | |||||
| Stages I, II | 26 | 74.3 | 19 | 54.3 | 0.081 |
| Stages III, IV | 9 | 25.7 | 16 | 45.7 | |
*P<0.05.
Tumour invasion of mucosa (m), submucosa (sm), muscularis propria (mp), subserosa (ss), penetration of serosa (se), and invasion of adjacent strucures (si).
Discussion
We recently reported that PICT1 is a novel oncogene that regulates the MDM2-TP53 pathway (Sasaki et al, 2011). The present study disclosed the function of PICT1 in gastric cancer, the most common malignant tumour in Japan. As expected, inhibition of PICT1 in gastric cancer cells resulted in wild-type TP53 accumulation and led to TP53-mediated cell cycle arrest and apoptosis. One of the critical findings of the current study was that PICT1 deficiency induced RPL11 translocation out of the nucleolus in gastric cancer cells, as we previously demonstrated for Pict1-deficient ES cells. Thus, this is the first demonstration that PICT1 regulates the nucleolar localisation of RPL11 in cancer cells and promotes TP53 accumulation, thereby reducing cancer progression.
As for the clinical usefulness of PICT1 expression in gastric cancer cases, we clarified that high expression of PICT1 correlated with a poorer prognosis in gastric cancer cases with wild-type TP53. We previously reported that PICT1 high expression levels indicated a poor prognosis in CRC and ESCC cases not only in TP53 wild-type cases but in all cases. On the other hand, our analysis of gastric cancer cases showed the prognostic significance of PICT1 expression but only in the TP53 wild-type group. It should be noted that our study patients with wild-type TP53 tumours had a significantly better prognosis than those with mutant TP53. As such, this TP53 status-related prognostic difference may explain this discrepancy. About 30% of human gastric cancers harbour TP53 mutations (Fenoglio-Preiser et al, 2003). Therefore, in 70% of gastric cancer cases with wild-type TP53, PICT1 expression status might determine the clinical outcome.
Activation of TP53 is induced by many types of cellular stress. PICT1 binds to ribosomal protein RPL11, leading to its localisation in the nucleolus. Reduction in PICT1 levels in response to nucleolar stress in particular releases RPL11 from the nucleolus followed by its translocation to the nucleoplasm. Serum depletion and some anticancer drugs, such as low-dose actinomycin D, mycophenolic acid (MPA), 5 fluorouracil, and camptothecin, are known to induce nucleolar stress, which is also referred to as ribosomal stress because of the inhibition of ribosomal biogenesis (Sun et al, 2008; Boulon et al, 2010). In this study, apoptosis was significantly induced in PICT1-deficienct cells following treatment with camptothecin and cisplatin (Figure 2D). In oligodendroglioma, loss of heterozygosity (LOH) of 19q13 was associated with better disease-free survival after chemotherapy (Cairncross et al, 1998; Smith et al, 1999; Mariani et al, 2006). These results indicate that gastric cancer patients having low PICT1 expression levels may retain higher chemotherapeutic susceptibility.
In summary, the level of PICT1 expression regulates the MDM2-TP53 pathway via RPL11 in gastric cancer and may be a novel prognostic factor in gastric cancer patients with wild-type TP53 tumours. Therefore, inhibition of PICT1 expression or interfering with the interaction between PICT1 and RPL11 might provide useful strategies for treating gastric cancer.
Acknowledgments
We thank T Shimooka and M Kasagi for their technical assistance. We also thank H Miyoshi (RIKEN BioResource Center) for providing lentivirus vector plasmid DNA, and Y Matsuzaki from the Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University for her technical support for sequence analyses. This work was supported in part by the following grants and foundations: CREST; Japan Science and Technology Agency (JST); Japan Society for the Promotion of Science (JSPS); Grant-in-Aid for Scientific Research, grant numbers 20390360, 20591547, 20790960, 21591644, 21791295, 21791297, 215921014, and 21679006; the Funding Program for Next Generation World-Leading Researchers (LS094); and NEDO (New Energy and Industrial Technology Development Organization).
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483 (7391:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40 (2:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90 (19:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D'Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39 (Database issue:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75 (4:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25 (13:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio-Preiser CM, Wang J, Stemmermann GN, Noffsinger A. TP53 and gastric carcinoma: a review. Hum Mutat. 2003;21 (3:258–270. doi: 10.1002/humu.10180. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20 (21:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387 (6630:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Inoue-Narita T, Hamada K, Sasaki T, Hatakeyama S, Fujita S, Kawahara K, Sasaki M, Kishimoto H, Eguchi S, Kojima I, Beermann F, Kimura T, Osawa M, Itami S, Mak TW, Nakano T, Manabe M, Suzuki A. Pten deficiency in melanocytes results in resistance to hair graying and susceptibility to carcinogen-induced melanomagenesis. Cancer Res. 2008;68 (14:5760–5768. doi: 10.1158/0008-5472.CAN-08-0889. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387 (6630:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Mariani L, Deiana G, Vassella E, Fathi AR, Murtin C, Arnold M, Vajtai I, Weis J, Siegenthaler P, Schobesberger M, Reinert MM. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J Clin Oncol. 2006;24 (29:4758–4763. doi: 10.1200/JCO.2006.05.9238. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80 (2:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Okahara F, Ikawa H, Kanaho Y, Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J Biol Chem. 2004;279 (44:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- Okahara F, Itoh K, Ebihara M, Kobayashi M, Maruyama H, Kanaho Y, Maehama T. Production of research-grade antibody by in vivo electroporation of DNA-encoding target protein. Anal Biochem. 2005;336 (1:138–140. doi: 10.1016/j.ab.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Okahara F, Itoh K, Nakagawara A, Murakami M, Kanaho Y, Maehama T. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol Biol Cell. 2006;17 (11:4888–4895. doi: 10.1091/mbc.E06-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, Hikasa H, Horie Y, Yamashita T, Kamijo T, Zhang Y, Zhu Y, Prives C, Nakano T, Mak TW, Sasaki T, Maehama T, Mori M, Suzuki A. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17 (8:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O'Fallon J, Yates A, Feuerstein BG, Burger PC, Scheithauer BW, Jenkins RB. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18 (28:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102 (43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem. 2008;283 (18:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6 (12:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, Piazuelo MB, Correa P, Washington MK, El-Rifai W, Peek RM, Zaika A. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139 (4:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim JH, Kim YJ, Ko JH, Cho YE, Kim SM, Kim JY, Lee S, Park JH. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ. 2007;14 (11:1872–1879. doi: 10.1038/sj.cdd.4402204. [DOI] [PubMed] [Google Scholar]
- Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T, Kuwano H, Nakayama KI, Mori M. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69 (9:3788–3794. doi: 10.1158/0008-5472.CAN-08-2846. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35 (3:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.