Fig. 2.
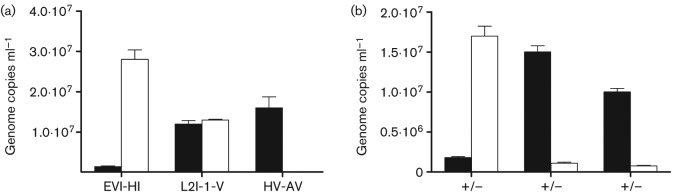
Enrichment efficiency of C. trachomatis serovar L2, as a factor of surface-reactive mAbs. qPCR of CT604 was used to infer bacterial enrichment efficiency based on chlamydial genome abundance. The white bars indicate genome copy numbers in the eluate of an IMS reaction, while the black bars indicate genome copies in the flow-through. (a) A laboratory-generated swab sample was harvested using antibodies to chlamydial LPS (EVI-HI), serovar L2 MOMP (L2-I-V) or to a serologically different MOMP (HV-AV) and processed through the IMS protocol. (b) Successful IMS only occurred in the presence of both primary and secondary antibodies. +/+, Recovery using both primary and secondary antibodies; +/−, no secondary antibody; –/+ indicates no primary antibody. This test was conducted using EVI-HI primary antibody.
