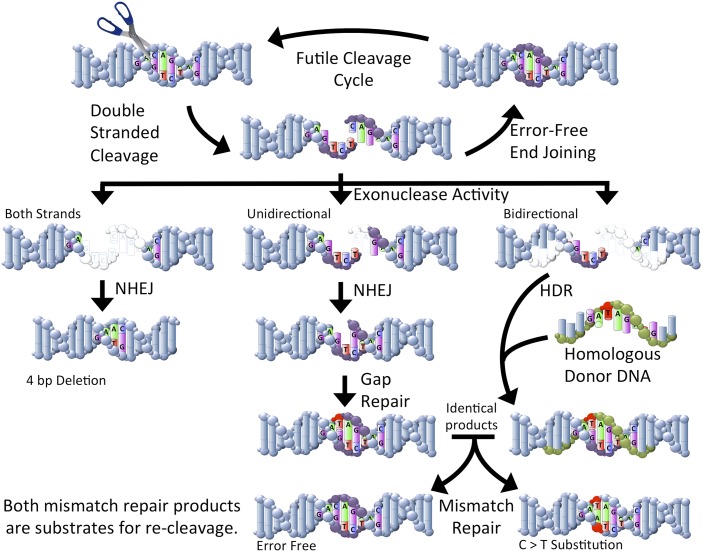
Fig. 1.
Simplified representation of the repair of double-strand breaks. Sequence-specific endonucleases can be designed to cleave genome targets (represented by scissors). In general, cleaved sites can be rejoined to correct the double-strand break. This cycle can continue until the nuclease is no longer present or until an error occurs during repair that prevents recleavage. Exonuclease activity can truncate the free ends (bases that have been removed are shown in white). When both strands are truncated, deletions can be produced by nonhomologous end-joining (NHEJ). The change in local spacing produces a sequence that cannot be recleaved (Left pathway). If exonuclease activity removes bases from one strand, a gap can be created after NHEJ. The missing bases can be replaced by polymerase activity (Center pathway). Although this process generally proceeds with high fidelity, errors can occur (C > T error shown in red). If homologous DNA is present (shown in green), homology-directed repair (HDR) can result in the introduction of specific modifications (C > T modification shown in red). The result shown here (Right pathway) produces a product that is identical to the error shown for gap repair. After further processing through mismatch repair, the C > T substitution can be corrected or can retain the substitution. In either case, the resulting products may be recleaved. Larger modifications can be made than shown here. In addition, larger repair templates can be used to facilitate homologous recombination.