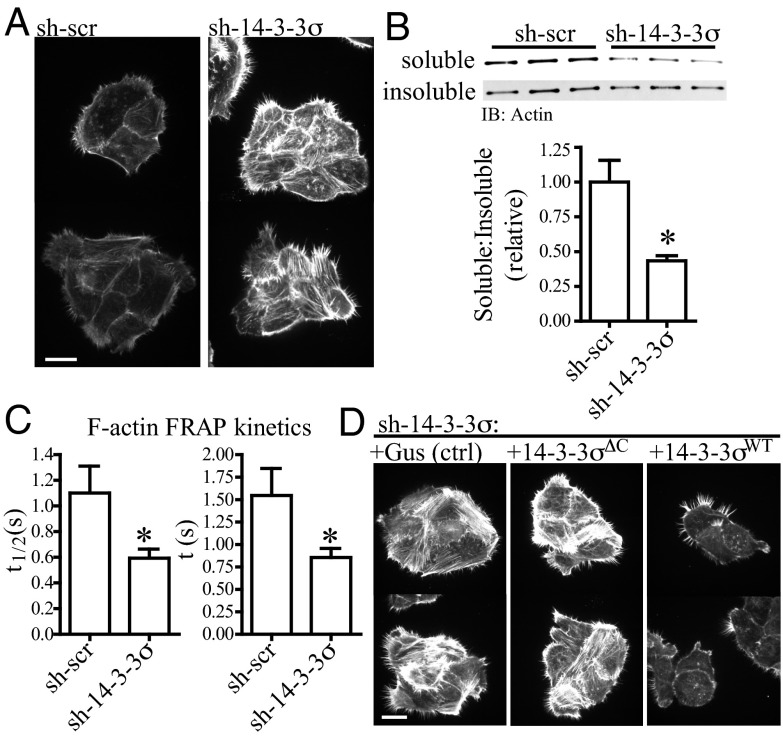
Fig. 2.
Protein 14-3-3σ regulates actin-polymerization kinetics in a ligand-binding mechanism. (A) Phalloidin immunofluorescence, showing T4-2 cells with reduced 14-3-3σ (sh-14-3-3σ) have deregulated F-actin polymerization relative to control cells (sh-scr). Images were acquired in the same focal plane using cells grown in adjacent wells of a chamber slide. (Scale bar: 20 μm.) (B) sh-14-3-3σ cells have decreased actin solubility. Cells were fractionated into 1% Triton X-100–soluble and –insoluble pools, and the ratio of soluble to insoluble actin was measured and quantified by Western blot. Error bars represent the SEM, and statistical significance was calculated using a two-tailed Student t test (*P < 0.05). (C) sh-14-3-3σ cells have increased actin turnover kinetics. F-actin dynamics in cells expressing LifeAct-mCherry were measured using FRAP. The kinetic parameter describing the rate of half (t1/2) or full (t) plateau fluorescence recovery was collected for n > 30 individual cells; lower values in sh-14-3-3 cells indicates a more rapid rate of fluorescence recovery and, thus, increased kinetics. Error bars represent the SEM, and statistical significance was calculated using a two-tailed Student t test (*P < 0.05). (D) Phalloidin immunofluorescence, showing the increased actin polymerization in sh-14-3-3σ cells can be rescued with forced expression of 14-3-3σWT but not an irrelevant gene (Gus) or the mutant 14-3-3σΔC. Images were acquired in the same focal plane using cells grown in adjacent wells of a chamber slide. (Scale bar: 20 μm.)