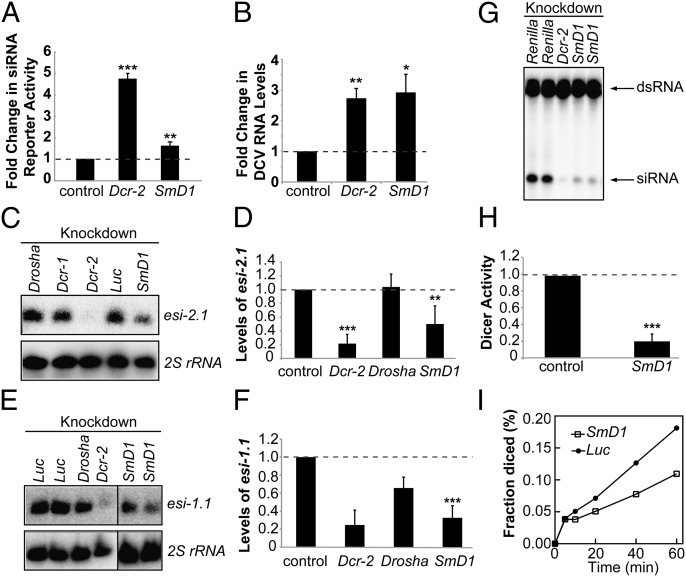
Fig. 1.
Loss of SmD1 compromises RNAi and siRNA biogenesis. (A) S2 cells were transfected with Dcr-2, SmD1, or control (LacZ) dsRNA together with expression constructs for a Renilla luciferase reporter containing complementary binding sites for esi-2.1 and a firefly luciferase control reporter. Loss of RNAi was measured as derepression of the luciferase reporter activity (n ≥ 3). (B) S2 cells treated with various dsRNAs (below) were infected with DCV. DCV RNA levels were measured by RT-qPCR (n ≥ 3). (C) Northern blotting to measure levels of the endogenous siRNA esi-2.1 or 2S rRNA (loading control) in various dsRNA-treated S2 cells. (D) Quantification of esi-2.1 levels (n = 4) normalized against 2S rRNA levels and compared with controls. (E) Northern blotting to measure esi-1.1 and 2S rRNA levels in various knockdown cells. (F) Quantification of esi-1.1 levels. Dcr-2 and Drosha, n = 2; SmD1, n = 4. (G) Cytoplasmic lysates from S2 cells treated with various dsRNAs were incubated with radiolabeled dsRNA substrate to generate 21-nt siRNAs. RNAs were extracted and resolved by denaturing urea-PAGE. (H) The amount of siRNAs is quantified and normalized to controls (n = 3). (I) Kinetic analysis of Dcr-2 activity using lysates from control or SmD1 knockdown cells. Fraction of dsRNA substrate processed by Dcr-2 at various time points is shown. Data are shown as mean + SD in A, D, F, and H or mean + SEM in B. *P < 0.05, **P < 0.01, ***P < 0.001. Unless noted otherwise, dsRNAs against Renilla luciferase or firefly luciferase serve as controls.