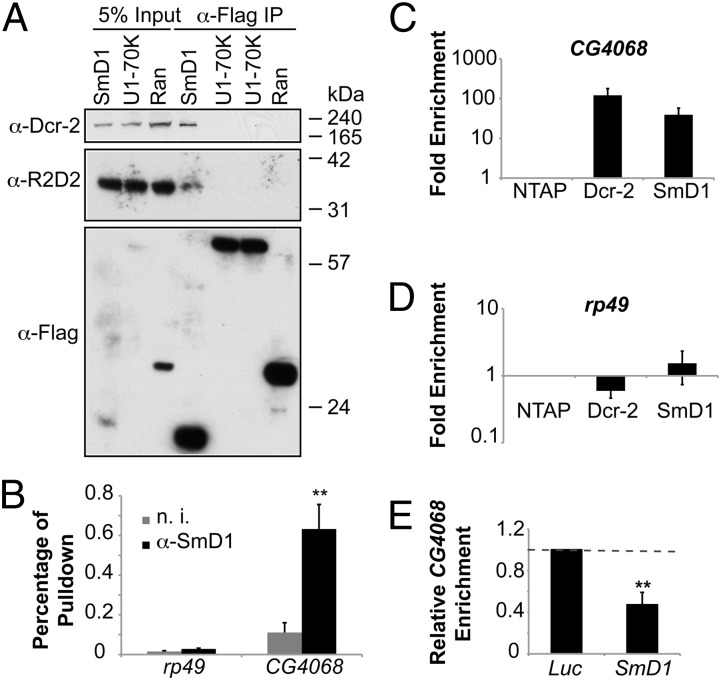
Fig. 2.
SmD1 interacts with components of the siRNA biogenesis machinery. (A) Lysates from S2 cells expressing Flag-tagged SmD1 or control proteins (U1-70K or Ran) were immunoprecipitated with anti-Flag antibody. Identical copies of input and immunoprecipitated material were subject to immunoblotting using various antibodies (left). (B) Total RNA was extracted from immunopurified SmD1 complexes and subject to RT-qPCR to measure levels of CG4068 (n = 4) or the control mRNA rp49. Samples recovered from a parallel immunoprecipitation using nonimmune serum (n. i.) serve as negative control. (C and D) Total RNA was extracted from immunopurified TAP-tagged Dcr-2 or SmD1 complexes and subject to RT-qPCR to measure levels of the esi-2.1 precursor transcript CG4068 (C) or the control mRNA rp49 (D). Fold enrichment (n = 3) compared with control samples (from cells expressing the TAP tag only) is shown on a log scale. (E) S2 cells stably expressing TAP–Dcr-2 were treated with control (Luc) or SmD1 dsRNAs. Relative enrichment of CG4068 in purified TAP–Dcr-2 complexes was measured by RT-qPCR (n = 3). Data are shown as mean + SD in E or mean + SEM in B, C, and D. **P < 0.01.