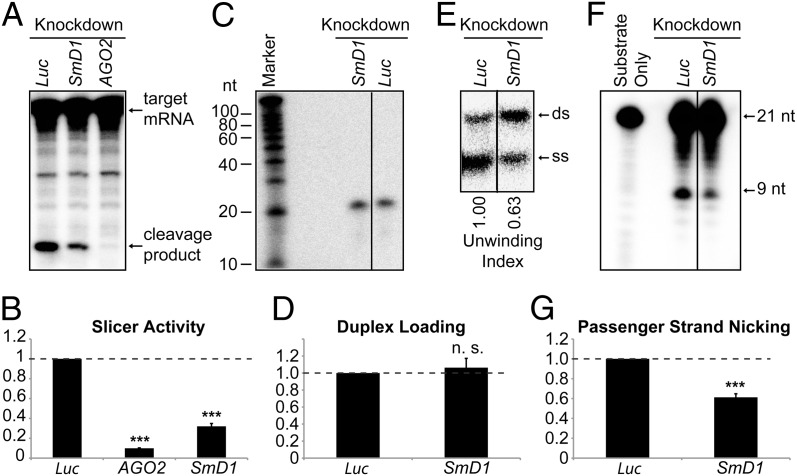
Fig. 3.
Loss of SmD1 abolishes siRISC assembly. (A) Cytoplasmic lysates from various knockdown cells (Top) were incubated with a synthetic siRNA duplex. The slicer activity of the resulting siRISC was measured as cleavage of the cap-radiolabeled target mRNA. (B) The slicer activity of the siRISC assembled in lysates from various knockdown cells (Bottom) was quantified (n ≥ 3). (C) Minimal RISC was immunopurified from Flag–AGO2 cell lysates using Flag-agarose beads. Purified minimal RISC was then incubated with lysates from control or SmD1-knockdown cells in the presence of an siRNA duplex radiolabeled on the guide strand. AGO2-bound RNAs were extracted from the beads and resolved by denaturing urea-PAGE. (D) siRNA duplex loading was quantified as the amount of radiolabeled siRNA guide strand recovered from the AGO2 siRISC assembled in lysates from various knockdown cells (n = 3). (E) siRNA duplex unwinding was measured as in C except that an excess of unlabeled guide siRNA strand was added to the reaction and AGO2-associated RNAs were resolved by native PAGE to separate ds-siRNAs and ss-siRNAs. The unwinding index was calculated and is shown at the bottom. A representative image from two independent experiments is shown. (F) Passenger-strand nicking was measured as described in C except that the siRNA duplex was radiolabeled on the passenger strand and an excess of 2′-O-methylated oligo complementary to the passenger strand was added. RNAs were extracted from the reaction mixture and resolved by denaturing urea-PAGE. (G) The passenger-strand nicking activity of lysates from various knockdown cells (Bottom) was quantified. Data are shown as mean + SEM in B, D, and G. ***P < 0.001; n.s., nonsignificant.