Fig. 5.
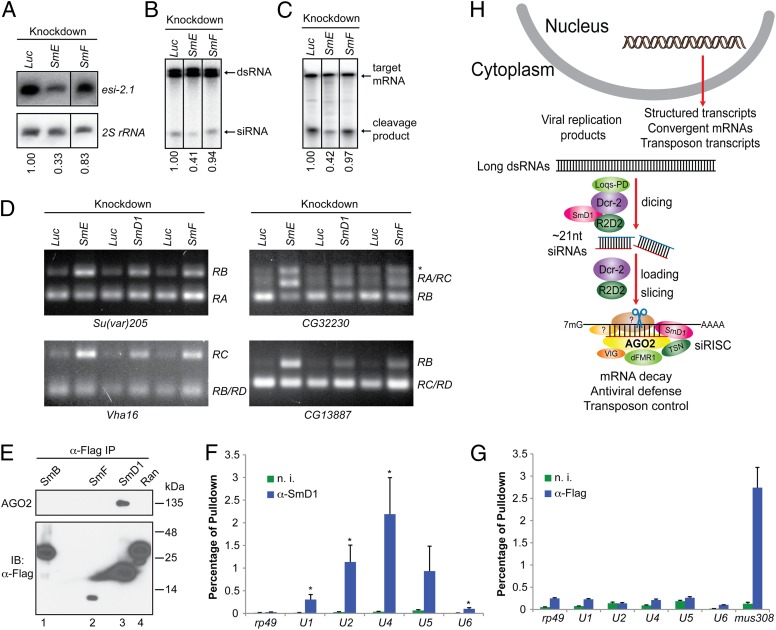
RNAi and splicing machineries are physically and functionally distinct. (A) S2 cells were treated with various dsRNAs. Levels of esi-2.1 and the control 2S rRNA were measured by Northern blotting. (B and C) Cytoplasmic lysates were assayed for (B) dsRNA processing by dicer assays and (C) mRNA cleavage by slicer assays. In A–C, relative levels of the indicated product are shown beneath the lanes. Representative images from two independent experiments are shown. (D) Splicing activity was assessed in various knockdown cells (Top) by examining splicing patterns of the indicated mRNAs (Bottom) by RT-PCR. The splice variants are shown on the right. The asterisk indicates an unannotated splice variant. (E) Lysates from S2 cells expressing various Flag-tagged proteins were immunoprecipitated using anti-Flag antibody. The immunoprecipitates were analyzed by immunoblotting with antibodies against AGO2 and Flag. (F and G) Total RNA was extracted from immunopurified SmD1 (F) or Flag–AGO2 complexes (G). Levels of various snRNAs or the control mRNAs mus308 and rp49 were measured by RT-qPCR. Results are the mean + SEM of n = 4; *P < 0.05. Samples recovered from a parallel immunoprecipitation using nonimmune serum (n. i.) serve as negative control. (H) A schematic depicting the proposed roles of SmD1 in the initiation (siRNA biogenesis) and effector (siRISC assembly and/or mRNA cleavage) steps of RNAi.