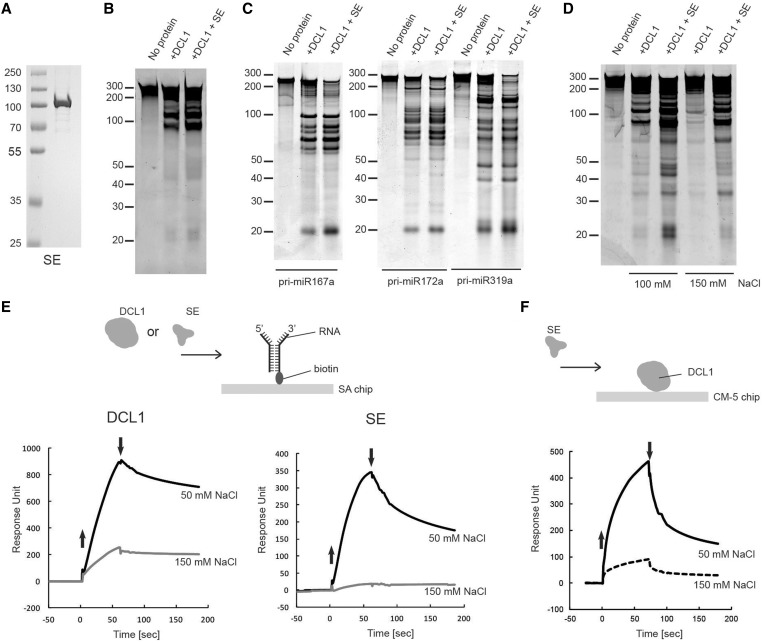
Figure 2.
Effect of SE on pri-miRNA processing. (A) Purified recombinant SE protein. Coomassie blue stained SDS–PAGE of purified SE expressed in a baculovirus/insect cell expression system. (B) Effect of SE on pri-miR167b processing by DCL1. Pri-miR167b (150 nM) was incubated with DCL1 (50 nM) and SE (240 nM) in a reaction buffer [20 mM Tris–HCl (pH 7.0), 50 mM NaCl, 5 mM MgCl2 and 1 mM ATP] at 37°C for 20 min. The RNA was fractionated on 12% polyacrylamide/7.5 M urea denaturing gel and visualized by SYBR Gold staining. (C) Effect of SE using three different pri-miRNA substrates. The reaction was done as in (B) with the indicated pri-miRNA substrates (150 nM). The RNA was fractionated and visualized as in (B). (D) Effect of SE on DCL1 at different NaCl concentrations. The reaction was done as in (B) with pri-miR167b, and the RNA was fractionated and visualized as in (B). (E) SPR analysis of interaction of DCL1 and SE with RNA. In all, 240 RU of biotin-labeled forked-RNA consisting of a 40-base pairs dsRNA region with a forked structure on one end that consist of a 20-base ssRNA on both strands was immobilized on the SA-coated sensor chip as shown in the schematic diagram, and DCL1 or SE was introduced for 70 s at a concentrations of 42 nM, in a buffer containing 20 mM Tris–HCl (pH 7.0), 2 mM β-ME, and either 50 mM or 150 mM NaCl. The up and down arrows indicate the start and end of protein injections, respectively. (F) SPR analysis of interaction of SE with DCL1. 1750 RU of DCL1 was immobilized via its amine group on the CM-5 sensor chip, and SE was injected for 70 s at a concentration of 20 nM in a buffer containing 20 mM Tris–HCl (pH 7.0), 2 mM β-ME, 0.005% surfactant P20 and 50 mM or 150 mM NaCl. The up and down arrows indicate the start and end of protein injections, respectively.