Figure 1.
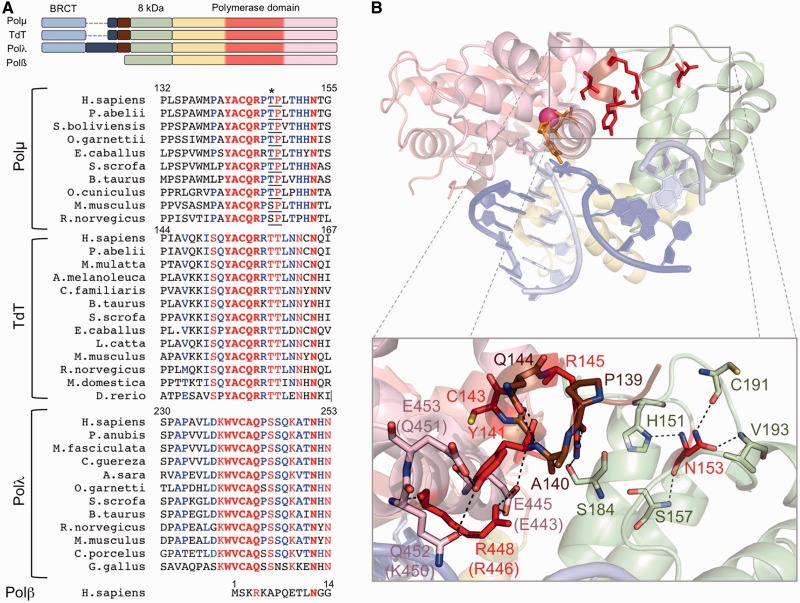
The brooch: amino acid conservation and implications for the closing of the polymerase core. (A) Top: schemes showing the domain organization of the human PolXs. The subdomains in the polymerase domain are coloured as follows: fingers in yellow, palm in red and thumb in pink. Bottom: multiple sequence alignment of Polλs, Polµs and TdTs of different species and human Polβ. Invariant residues inside each group are shown in red, conserved residues in blue. The brooch and the invariant asparagine equivalent to Polµ Asn153 are highlighted in bold. Asterisk indicates a Cdk phosphorylation residue at the minimal consensus sequence (TP/SP; underlined). (B) Cartoon representation of the murine Polµ ternary complex with the domains coloured as in (A). Labels of some residues are given for both mouse and human (in parentheses) counterparts, as they differ. The DNA substrate is shown in sticks, dark blue for the template strand and light blue for the primer and downstream strands. One nucleotide of the template strand has been eliminated to mimic a NHEJ intermediate formed by two 3′-protruding substrates. Residues from the brooch are shown in brown, unless those subjected to mutagenesis, which are shown in red sticks; residues contacting them directly are shown in the colour of the subdomain they belong to.