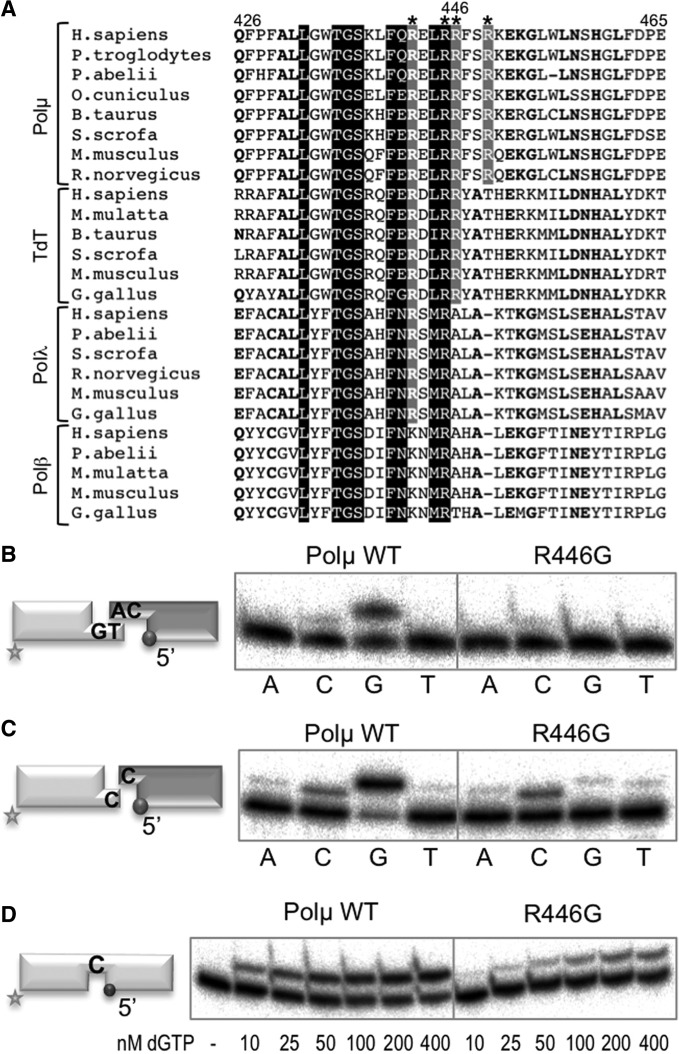
Figure 4.
Mutation of the interacting thumb residue Arg446 mimics disruption of the brooch. (A) Multiple sequence alignment showing the region corresponding to helix N in the four X family polymerases from different species. Numbers indicate the residues in the human Polµ sequence. The four conserved arginines (Arg444, Arg447, Arg448 and Arg451 in mouse Polµ) oriented towards the negatively charged phosphate backbone of the template strand are boxed in grey or black and indicated with an asterisk. Arg446 in human Polµ is numbered. (B) NHEJ reactions were performed for 30 min at 30°C with 400 nM of the indicated proteins and using two sets of substrates: the labelled substrate contained 1TG and 1D-NHEJ and the cold one 2AG and 2D-NHEJ. Each of the four ddNTPs (100 µM) was added in the presence of 2.5 mM MgCl2. (C) NHEJ reactions were performed as in (B) but using non-complementary DNA substrates, the labelled one containing 1C and 1D-NHEJ, and the cold one containing 1C and 2D-NHEJ. (D) Gap-filling reactions were performed for 30 min at 30°C with the indicated proteins (100 nM) using a gapped substrate containing SP1C, T13C and DG1-P. dGTP was added separately at the indicated concentrations in the presence of 2.5 mM MgCl2.