Table 2.
Multi-profile alignment of the glycomic dataset with and without using a GP prior
| Performance measure | Binning without GP prior |
Clustering without GP prior |
Binning with GP prior |
Clustering with GP prior |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 | |
| Precision | 0.957 | 0.960 | 0.964 | 0.963 | 0.964 | 0.980 | 0.979 | 0.980 | 0.976 | 0.977 | 0.978 | 0.974 | 0.980 | 0.979 | 0.980 | 0.980 |
| (0.007) | (0.008) | (0.007) | (0.006) | (0.007) | (0.004) | (0.004) | (0.002) | (0.003) | (0.003) | (0.004) | (0.006) | (0.003) | (0.003) | (0.002) | (0.003) | |
| Recall | 0.766 | 0.783 | 0.770 | 0.780 | 0.796 | 0.906 | 0.910 | 0.913 | 0.848 | 0.856 | 0.830 | 0.837 | 0.904 | 0.904 | 0.907 | 0.908 |
| (0.138) | (0.122) | (0.147) | (0.154) | (0.138) | (0.094) | (0.099) | (0.092) | (0.151) | (0.141) | (0.160) | (0.156) | (0.098) | (0.099) | (0.095) | (0.096) | |
Note: Precision and recall are compared between cases where G chromatograms (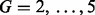 ) are derived either by binning along m/z or using the chromatographic clustering procedure.
) are derived either by binning along m/z or using the chromatographic clustering procedure.
