Figure 6. Acetylation modulates NS1 transcriptional activities.
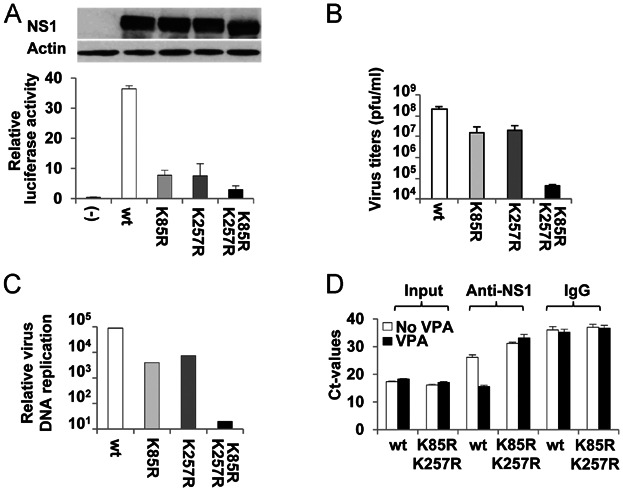
- The K85R and K257R mutations decrease NS1-dependent transactivation of the P38 promoter. HeLa cells were co-transfected with pGL3-H-1PV-P38, pRL-TK luciferase and the pCDNA4/TO plasmid, either empty (−) or expressing a wild-type NS1 (wt) or an acetylation-defective mutant form of this protein. 48 h post-transfection, the cells were processed for the firefly Dual-Luciferase Assay. Western blot analysis was performed with total cell lysates from these cultures in order to ascertain equal NS1 protein levels.Source data is available for this figure in the Supporting Information.
- Mutation of both NS1 acetylation sites strongly impairs H-1PV production. NB324K cells were infected with the same amount (100 viral genome/cell) of either H-1PV wild-type (wt) or acetylation-defective H-1PV-K85R, H-1PV-K257R, or H-1PV-K85R-K257R for a 6-day round of virus amplification. Production of progeny viral particles was determined by plaque assay. Average values from a representative experiment performed in triplicate are shown with standard deviation bars.
- Mutation of both NS1 acetylation sites strongly reduces H-1PV DNA replication. Viral DNA from HeLa cells infected with the indicated virus was analysed by Southern blot analysis and quantified with a PhosphorImager.
- Mutation of the NS1 acetylation sites strongly impairs the capacity of the protein to bind to the H-1PV P38 promoter. HeLa cells were infected with either H-1PV wild-type (wt) or with the H-1PV-NS1-K85R-K257R mutant and grown with (black bars) or without (white bars) VPA (1 mM) for 24 h before processing for ChIP assays as described in Fig 3E.
