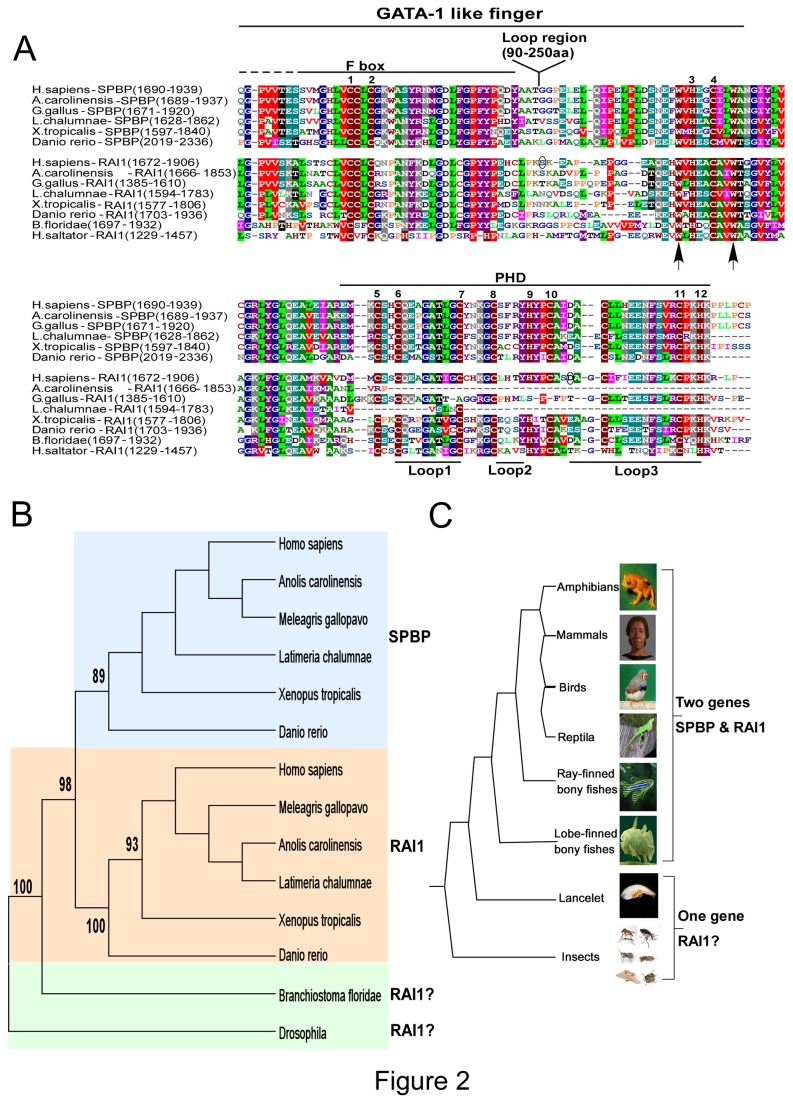
Figure 2. The ePHD/ADD domains of SPBP and RAI1 are conserved in evolution.
(A) Alignment of the putative ePHD/ADD domain of SPBP and RAI1 in mammalia, reptiles, birds, amphibians, fishes and insect. Cysteine and histidine residues that may serve as zinc-ligands are indicated by numbers above the alignment. The long loop region between zinc ligands 2 and 3 in all species is excluded from the alignment, but indicated above. The regions encompassing the putative GATA-1 like finger and the PHD finger are indicated above, while the Loop1, Loop2 and Loop3 indicated below are loop structures generally found in PHD fingers. The conserved Tryptophan units used to select blasted sequences are indicated by arrows, while amino acids probably involved in Smith-Magenis syndrome are encircled. Two of the RAI1 sequences (A.carolinensis and L.chalumnae) are truncated in the C-terminal end probably due to sequencing errors. The threshold for shading in manually refined Clustal W sequence alignment was set to 50% using BLOSUM 62 scoring matrix. (B) Phylogenetic tree of the ePHD/ADD domains of SPBP and RAI1 proteins. The phylogenetic tree was constructed by Mega5 software using the maximum likelihood method. Numbers in the branch represents the bootstrap values. Background color coding is used to represent species which possess SPBP (blue), RAI1 (pink) and uncharacterized SPBP/RAI1 like proteins (green). The phylogenetic tree is based on selected protein sequences from representative vertebrate organisms (e.g. Homo sapiens represent Mammalia). (C) The evolutionary tree of the ePHD/ADD domains of SPBP and RAI1 proteins. The alignment of all sequences is shown in Supporting information, Figure S1 and the protein sequence accession numbers are provided in the Supporting information, Table S3.