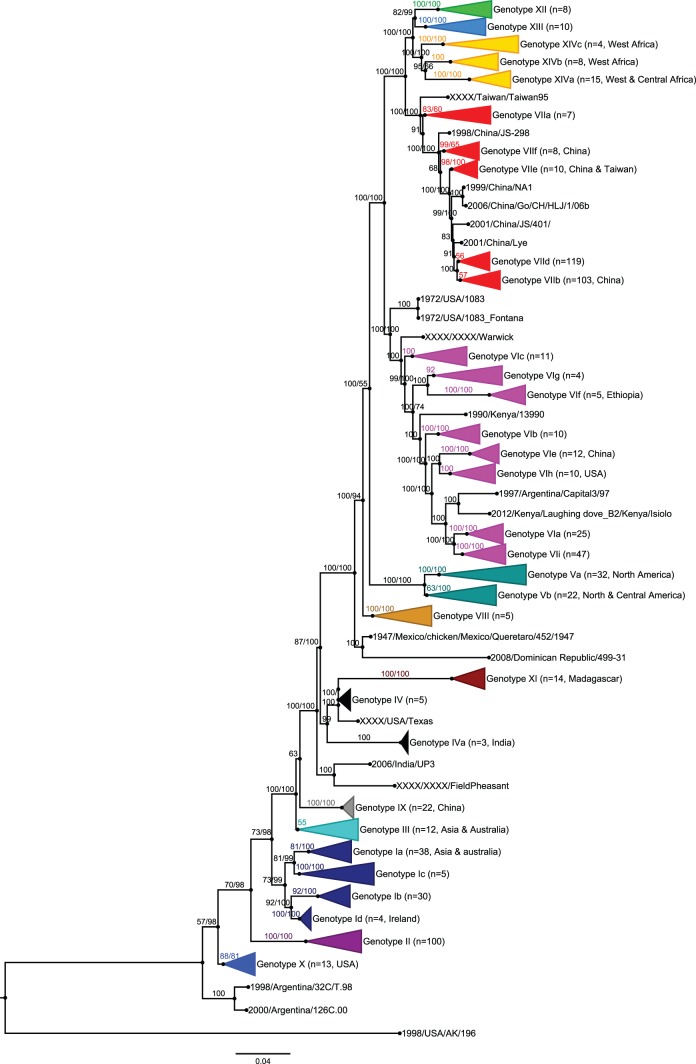
Figure 1. Phylogenetic analysis of 741 complete F nucleic acid sequences of NDV.
Trees were constructed using Bayesian inference with 16,806,000 iterations and 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 1681, and PSRF were between 0.99996 and 1.00084. A class 1 virus sequence was introduced as an outgroup. Consensus tree posterior probabilities are indicated on the branch (first number). A Bayesian inference based on 323 complete HN nucleic acid sequences was also done (all details in Figure S3A, Figure S5J and Figure S9R). Since the F and HN trees were superimposable, only the F tree is represented; where informative, branch support values for HN are indicated after the F posterior probabilities. Sequences from this study are grouped in genotypes Ib (2 strains), VIf (5 strains), XI (10 strains), XIVa (1 strains), XIVb (5 strains), and XIVc (1 strain). The complete list of the 741 sequences, the corresponding multiple sequence alignments, and the tree in Newick and in Figtree format can be found in Table S8, Table S7, Figure S5G, and Figure S9O, respectively.