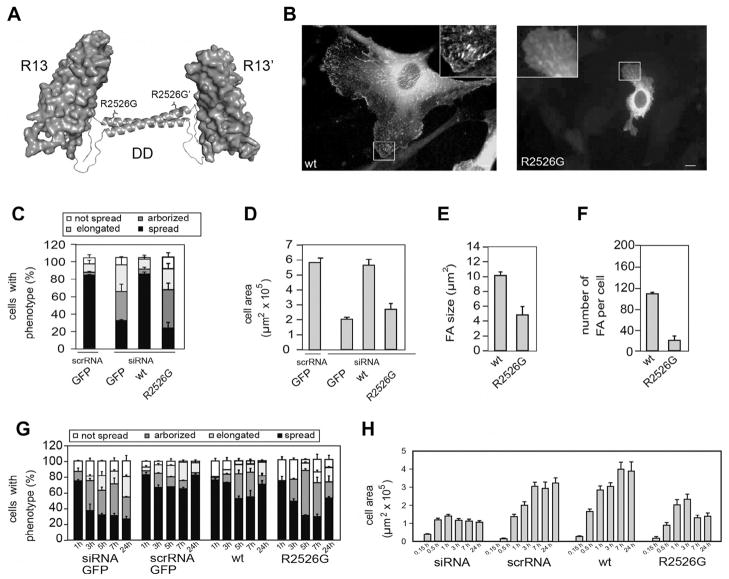
Fig. 2.
The ability of talin1 to support cell spreading and focal adhesion assembly is compromised by a R2526G mutation in the dimerization domain (DD). (A) The C-terminal actin-binding site of talin1 is comprised of rod domain R13 (a 5-helix bundle) and a C-terminal helix, the dimerization domain (DD). Mutation of R2526G abolishes dimerization of a R13-DD polypeptide and markedly reduces its affinity for filamentous actin (Gingras et al., 2008). (B) GFP-talin1 (left panel; WT) supported extensive cell spreading and was localized in abundant focal adhesions (FAs). In contrast, cells expressing the GFP-talin1 R2526G mutant were much less well spread and had far fewer FAs. (C) Cell morphology – cells were classified into four groups; not spread, elongated (cells 5× longer than wide), arborised (cells with >5 protrusions) or spread. (D) Cell area (E) FA size and (F) number per cell were quantified using ImageJ (NIH). Compared to cells expressing GFP-talin1, those expressing the GFP-talin1 R2526G mutant were more elongated or arborised, had a much reduced cell area, and FAs were reduced in number and size. (G, H) Time-course of cell spreading. Cells expressing GFP-talin1 R2526G managed to spread during the first 1 h after plating, but thereafter, the cells could not maintain a spread phenotype, and the spread cell area remained low.