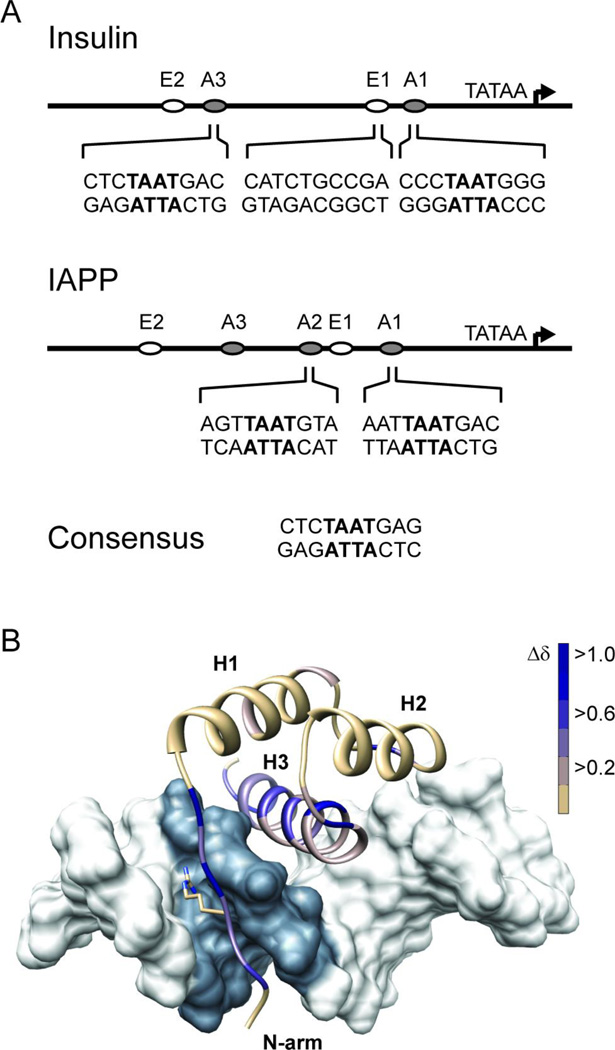
Figure 1.
Pdx1 recognizes A-box motifs common to the insulin and iapp gene promoters. (A) Schematic representations of the insulin and iapp gene promoters depict the relative location of A-box promoter elements, and the adjacent E-box elements, which are also involved in regulating transcription from these promoters. The nucleotide sequence in the neighborhood of the core TAAT element (denoted in bold) is shown for each of the four A-boxes investigated in this study. Pdx1 does not bind to E-boxes and so the insulin E1 sequence is used as a control for non-specific DNA binding in this study. (B) The DNA binding epitope of Pdx1 is confirmed by monitoring changes in the backbone amide chemical shift between the unbound and consensus-DNA bound states. Chemical shift changes (Δδ) are mapped as progression from tan to blue onto a ribbon-representation of Pdx1 in complex with duplex-DNA (molecular surface of pdb 2h1k, in which the core TAAT element is depicted in grey). Δδ represents the weighted change in amide 1H and 15N chemical shift for each residue and is calculated using Equation 1 (see Methods). Pdx1 secondary structural elements are labeled as H1 – H3 (α-helix-1,2,3 respectively) and the N-terminal arm is also labeled for reference. Also depicted is the side chain of Arg-150, which is shown in the crystallographic orientation.