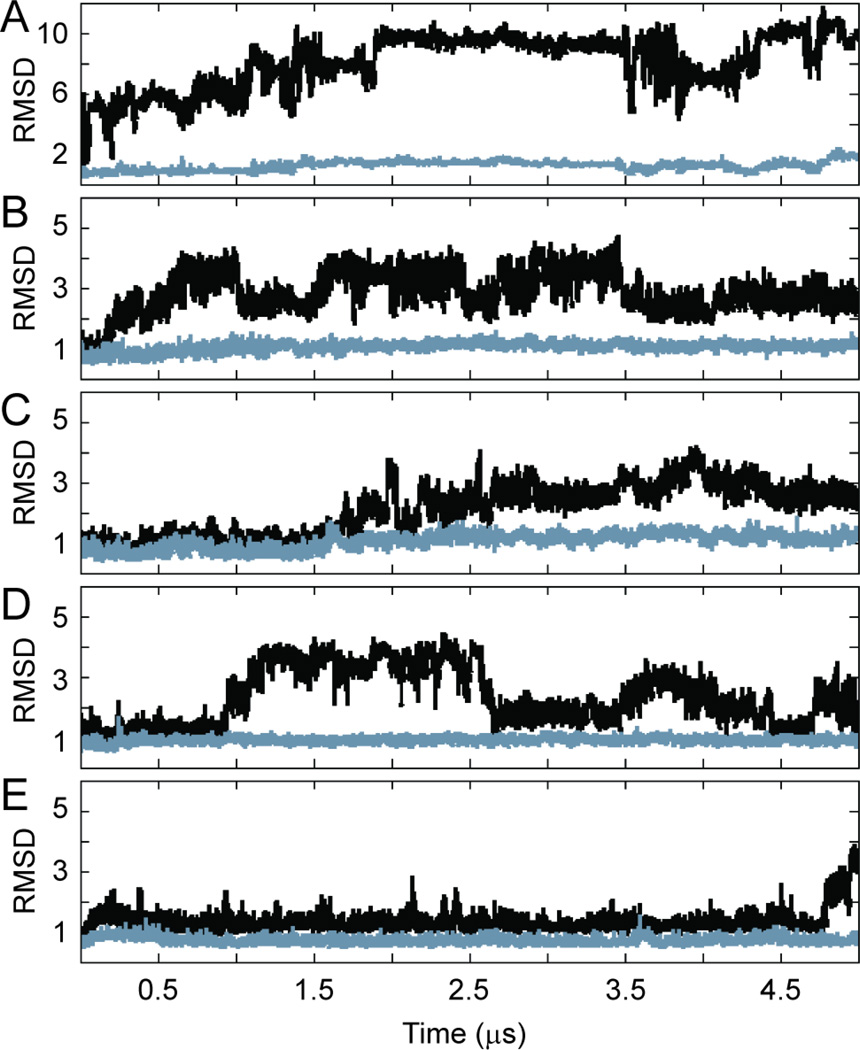
Figure 3.
Pdx1 root-mean-squared deviation (RMSD) from the crystal structure during the five-microsecond production molecular dynamics calculations performed on Anton. Results are shown for (A) apo-Pdx1, (B) Pdx1-DNACON(CTAATG), (C) Pdx1-DNA(ATAATG), (D) Pdx1-DNA(GTAATG), and (E) Pdx1-DNA(TTAATG). For each trajectory, the Pdx1 RMSD (Å) is shown for all backbone heavy atoms (black) and for the backbone heavy atoms of residues 155–199 only (grey). Data points are reported once per 200 ps of simulaton time.