Abstract
Developed to compliment the use of fluorescent proteins in live cell imaging, chemical tags enjoy the benefit of modular incorporation of organic fluorophores, opening the possibility of high photon output and special photophysical properties. However, the theoretical challenge in using chemical tags as opposed to fluorescent proteins for high-resolution imaging is background noise from unbound and/or non-specifically bound ligand-fluorophore. We envisioned we could overcome this limit by engineering fluorogenic trimethoprim-based chemical tags (TMP-tags) in which the fluorophore is quenched until binding with E. coli dihydrofolate reductase (eDHFR) tagged protein displaces the quencher. Thus, we began by building a non-fluorogenic, covalent TMP-tag based on a proximity-induced reaction known to achieve rapid and specific labeling both in vitro and inside of living cells. Here we take the final step and render the covalent TMP-tag fluorogenic. In brief, we designed a trimeric TMP-fluorophore-quencher molecule (TMP-Q-Atto520) with the quencher attached to a leaving group that, upon TMP binding to eDHFR, would be cleaved by a cysteine residue (Cys) installed just outside the binding pocket of eDHFR. We present the in vitro experiments showing that the eDHFR:L28C nucleophile cleaves the TMP-Q-Atto520 rapidly and efficiently, resulting in covalent labeling and remarkable fluorescence enhancement. Most significantly, while only our initial design, TMP-Q-Atto520 achieved the demanding goal of not only labeling highly abundant, localized intracellular proteins, but also less abundant, more dynamic cytoplasmic proteins. These results suggest that fluorogenic TMP-tag can significantly impact highresolution live cell imaging and further establish the potential of proximity-induced reactivity and organic chemistry more broadly as part of the growing toolbox for synthetic biology and cell engineering.
The past two decades have seen a transformation in cell biology brought about by the fluorescent proteins (FPs) as selective, genetic protein tags for live cell imaging.(1, 2) The original green fluorescent protein (GFP) from A. victoria is a 238 amino acid protein, which, upon folding, spontaneously forms a fluorescent chromophore by rearrangement and oxidation of Ser, Tyr, and Gly residues in the core of the eleven-stranded β-barrel.(3, 4) Since these original reports, naturally occurring and engineered FPs have been routinely used to observe the timing and location of protein expression in living cells, often providing significant mechanistic insight.(3, 5, 6) While FPs continue to be invaluable tools for cell biology, they have limitations for the increasingly sophisticated biophysical experiments necessary to make dynamic measurements of protein interactions critical to unraveling the molecular mechanism of cellular processes. Thus, chemical tags have been developed to provide an alternative for labeling intracellular proteins with modular organic fluorophores that have high photon outputs and other specialized functionalities.(7)
With chemical tags, rather than tagging the target protein with a FP, the protein is tagged with a polypeptide or protein receptor, which is subsequently modified with an organic fluorophore. The TMPtag, developed by Cornish and Sheetz, relies on the high-affinity interaction between E. coli dihydrofolate reductase (eDHFR) and the folate analog trimethoprim (TMP).(8, 9) In brief, the target protein is tagged with eDHFR, which binds TMP with high affinity (∼ 1 nM KD) and selectivity (affinities for mammalian DHFRs are KD > 1 μM).(10, 11) Organic fluorophores can be conjugated to TMP with only minor perturbation of the high-affinity binding to eDHFR. Among numerous chemical tags reported in the past decade, TMP-tag is one of few chemical tags that can label proteins with high efficiency and selectivity both in vitro and inside of live cells.(7) Significantly, the advantage of TMP-tag for highresolution imaging has been verified by several recent reports, including single-molecule (SM) imaging of spliceosome assembly in yeast cell extracts and super-resolution imaging of nucleosomes in live HeLa cells.(12, 13)
Nevertheless, one major challenge to realizing the potential of TMP-tag for high-resolution imaging is background from unbound and/or non-specifically bound fluorophore. Therefore, we set out to build a fluorogenic TMP-tag in which the fluorophore is switched on as TMP binds to eDHFR, minimizing the background fluorescence. Looking back into the rich history of fluorogenic probes, we found a number of viable designs for switch-on fluorophores. One widely used approach relies on fluorophores with protecting groups that are cleaved by an enzyme or light irradiation.(14, 15) The widely utilized Ca2+ sensors Fluo-3 and Rhod-2 represent a class of fluorogenic probes modulated via photon-induced electron transfer (PeT).(16) Other strategies include solvatochromes that are sensitive to the polarity of the microenvironment and molecular rotors that only fluoresce when intramolecular rotation is constrained.(17-19) Notably, the first chemical tag - the fluorogenic FlAsH tag reported in 1998 - utilizes fluorogenic biarsenical fluorophores hypothetically based on the molecular rotor or PeT concept.(20) Here we designed a fluorogenic TMP-tag by conjugating the TMP-fluorophore to a quencher that is cleaved when TMP binds to eDHFR. We choose the cleavable fluorophore/quencher strategy because: (1) it gives among the highest fluorescence enhancements; (2) the fluorescence enhancement hinges on the specific receptor/ligand binding; and (3) it allows for modular incorporation of virtually any organic fluorophore.
In the primary stage of achieving our goal of developing a fluorogenic TMP-tag, we first rendered the TMP-tag covalent via proximity-induced Michael addition.(21) In brief, we installed a unique Cys residue on eDHFR in position to react with a latent acrylamide electrophile added to the TMP-probe molecule. Exceeding our expectation, the first-generation covalent TMP-tag undergoes rapid, quantitative covalent labeling (in vitro t1/2 ∼ 50 min at 1 μM eDHFR and TMP-tag) and enables imaging of nuclear-localized eDHFR in live NIH3T3 cells. Nevertheless, the original covalent TMP-tag was not efficient enough to label cytoplasmic proteins that are less abundant and more dynamic, likely due to its modest reactivity. Therefore, by designing a shorter, optimized linker between TMP and the acrylamide electrophile as well as screening a panel of eDHFR variants, we recently engineered a second-generation covalent TMPtag that undergoes rapid, quantitative covalent labeling (in vitro t1/2 ∼ 8 min at 1 μM eDHFR and TMP-tag) and enables live cell imaging of several different cytoplasmic proteins.(22)
Here we further challenge the potential of proximity-induced reactivity by applying another classic type of reaction in organic chemistry, the SN2 reaction, to generate a fluorogenic TMP-tag suitable for live cell imaging (Figure 1). First, a tosylate electrophile and Cys nucleophile were selected as the reactive functional groups for modification of TMP and eDHFR, respectively. Using molecular modeling programs, we designed and synthesized a heterotrimeric TMP-tosylate-fluorophore with a quencher attached to the tosylate leaving group, with the Cys installed on the surface of eDHFR in position to displace the tosylate. Then the TMP analog was characterized in vitro and in live cells in order to determine the efficiency and selectivity of labeling. Finally, the utility of the fluorogenic TMP-tag was assessed by challenging it to label several different fusion proteins in live cells. Not only does this work provide a new fluorogenic tag for live cell imaging with high signal-to-background (S/B), but also it demonstrates that proximity-induced labeling is a viable tool for engineering cellular reagents.
Figure 1.
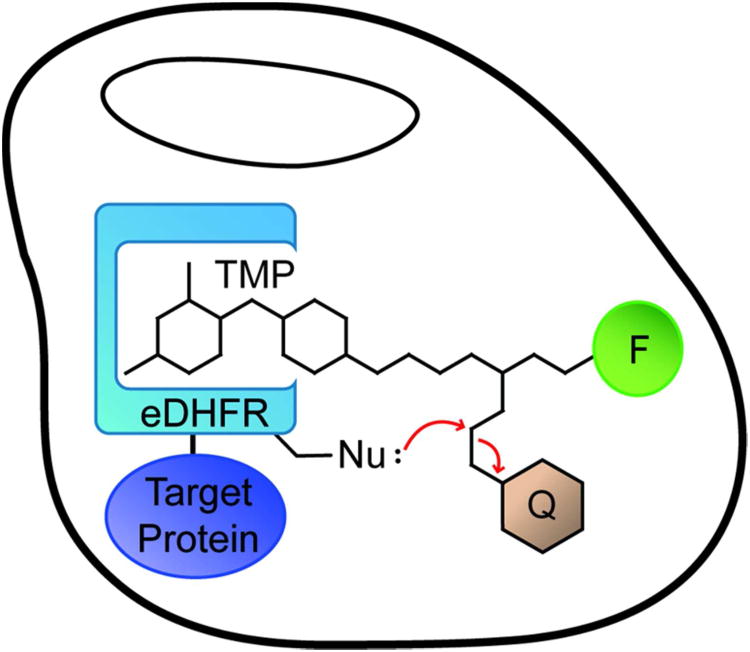
Cartoon of fluorogenic TMP-tag. Fluorogenic TMP-tag is based on the proximity-induced reaction between the E. coli dihydrofolate reductase (eDHFR) variant and the trimethoprim (TMP) – fluorophore (F) – quencher (Q) heterotrimer. The target protein (purple) is tagged with eDHFR (blue) and then labeled with TMP analogue. The high affinity interaction between TMP and eDHFR induces the SN2 reaction between a nucleophilic amino acid residue (Nu:) engineered on eDHFR and an electrophilic linker attached to the quencher as the leaving group.
Results and Disscussion
Design of a fluorogenic TMP-tag
We envisioned constructing the fluorogenic TMP-tag based on our covalent TMP-tag, centered on a proximity-induced SN2 reaction. Specifically, we designed a fluorogenic TMP-tag as a heterotrimeric molecule with the TMP, the fluorophore, and the quencher linked via an electrophilic leaving group. Upon binding to the eDHFR:Cys variant, the electrophilic leaving group is attacked by the Cys nucleophile on eDHFR, leading to displacement of the quencher along with the leaving group, producing a fluorescence enhancement (Figure 2a). To achieve live cell imaging with high S/B, we selected a fluorophore, quencher, and electrophile that would be cell permeable and well behaved inside of living cells.
Figure 2.
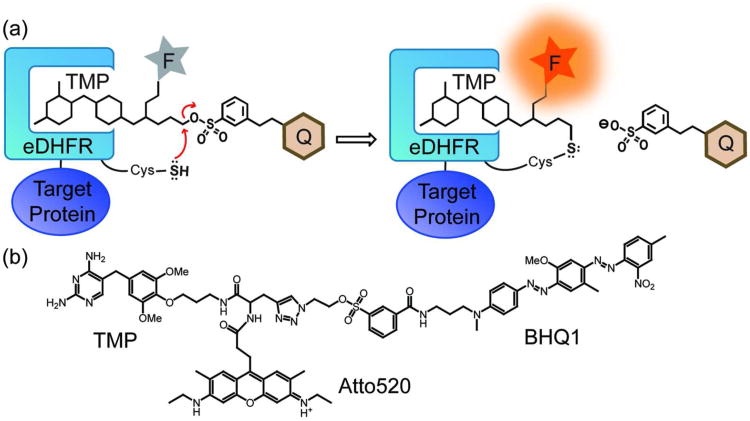
The fluorogenic TMP-tag. a) Cartoon of the fluorogenic TMP-tag, which centers a trimeric TMP-quencher (Q) - fluorophore (F) molecule to be cleaved in a proximity-induced SN2 reaction. Specifically, we selected the Cys side chain as the nucleophile and a tosylate linker as the electrophile. When TMP binds to eDHFR, the unique Cys near binding pocket would replace the tosylate leaving group attached to the quencher, and thus the probe is switched on. b) Structure of TMP-BHQ1-Atto520 (TMP-Q-Atto520), the first fluorogenic TMP-tag.
For the fluorophore, we chose Atto520 because it is a green dye (Ex: 516 nm; Em: 538 nm) with excitation and emission spectra similar to that of enhanced green fluorescent protein (EGFP) and is compatible with most fluorescence microscopes.(23) More importantly, Atto520 has been reported to have high photon output and photostability, verified by its application in super-resolution imaging in fixed cells.(24) Compared to other commonly used green fluorophores such as fluorescein, Alexa488 and Atto488, the unique positive charge on Atto520 is supposed to render it readily cell permeable, which is crucial to live cell imaging.
To achieve maximal quenching effect, the absorption spectrum of the quencher has to match the emission spectrum of the fluorophore. Thus we chose the black hole quencher 1 (BHQ1®, Biosearch Technologies) that shows broad and strong absorption across the green-yellow region peaking at 534 nm (ε = 38,000 M−1cm−1). The BHQ1 quencher is frequently used in nucleic acid detection technologies such as quantitative PCR and for molecular beacons, demonstrating high quenching efficiency with many green fluorophores.(25)
The key to our fluorogenic design is the choice and precise modification of the electrophilic leaving group. Specifically, the electrophilic functional group has to be reactive enough to facilitate rapid protein labeling upon TMP binding, yet resistant to the numerous nucleophilic biomolecules inside of live cells.(21, 26) Among various leaving groups commonly used in organic synthesis, we focused on the tosylates and mesylates because they potentially balance low reactivity towards endogenous cellular molecules with efficient proximity-induced reactivity when TMP binds to eDHFR.
Particularly, we found the quenched ligand-directed tosyl (Q-LDT) chemistry to be promising because it has been reported to have rapid proximity-induced in vitro reactivity with minimal non-specific hydrolysis.(27, 28) Furthermore, we anticipated we could significantly accelerate the labeling reaction by using the Cys nucleophile that is much stronger than the His nucleophile originally reported for the Q-LDT chemistry. Specifically, we built the TMP-quencher-fluorophore (TMP-Q-Atto520) molecule with the tosylate group 12 atoms away from the TMP para-methoxy group that locates at the opening of the TMP binding pocket. With this design the eDHFR/TMP interaction is minimally perturbed, yet the tosylate electrophile is positioned close to the surface of eDHFR to efficiently induce the SN2 reaction with the Cys nucleophile (Figure 2b).
Synthesis of TMP-Q-Atto520
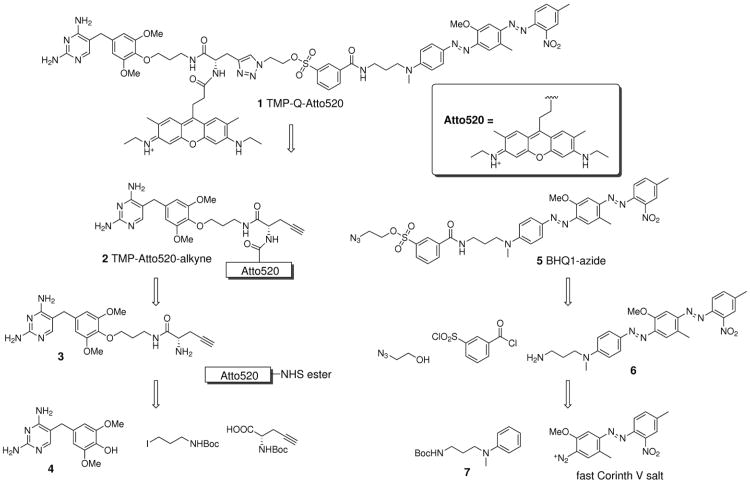
The synthetic design of the fluorogenic TMP-tag was planned around a tribranched core with 3 orthogonally reactive functional groups for coupling the TMP, the fluorophore, and the leaving group and quencher, respectively. Specifically, we used propargylglycine, taking the amino group and a carboxylic acid group to be attached to the fluorophore and the TMP, respectively, and the alkyne group for introduction of the quencher via a Cu(I) catalyzed azide-alkyne cycloaddition ‘click’ reaction.
As outlined in Scheme 1, the synthesis of the first half of the molecule, the TMP-Atto520-alkyne, started from the derivatization of TMP at the 4′-methoxy position, well established not to disrupt binding to eDHFR. The TMP-OH (intermediate 4) thus generated was then alkylated with commercially available tert-butyl-N-(3-iodopropyl)carbamate to produce TMP-C3-NHBoc. Then after removing the Boc group with trifluoroacetic acid (TFA), the TMP-C3-amine was coupled to N-Boc-propargylglycine under standard peptide coupling conditions, yielding TMP-NHBoc-alkyne. In the next step, the fluorophore Atto520-NHS ester was reacted with intermediate 3 in which the amino group was exposed by another TFA deprotection, producing intermediate 2. Notably, this synthetic route allowed us to introduce the fluorophore at a late stage in the synthesis, important not only because of fluorophore expense but also to allow for future synthesis of fluorogenic TMP-tags at different wavelengths.
Scheme 1.
Retro-synthetic design of TMP-Q-Atto520.
In parallel, the synthesis of the BHQ1 quencher was initiated with Boc protection of the commercially available N-(3-aminopropyl)-N-methylaniline. The product, intermediate 7, was then coupled to fast Corinth V salt in an azo coupling reaction following the patent US 7019129 B1.(29) With the Boc protective group removed by TFA, the BHQ1-amine (intermediate 6) was then derivatized with 3-(chlorosulfonyl)benzoyl chloride to produce BHQ1-tosyl chloride. In the next step BHQ1-tosyl chloride was reacted with excess 2-azidoethanol, catalyzed by 4-dimethylaminopyridine (DMAP), yielding intermediate 5.
Finally, the TMP-Atto520-alkyne (intermediate 2) and BHQ1-azide (intermediate 5) were conjugated via click chemistry under mild conditions (CuSO4·5H2O and ascorbic acid in anhydrous DMF). Importantly, the ascorbic acid kept the solution slightly acidic and thus minimized alkaline hydrolysis of the tosylate. The final product TMP-Q-Atto520 (molecule 1) was obtained in 2 percent overall yield from TMP or 3 percent overall yield from N-(3-aminopropyl)-N-methylaniline, both in five linear steps.
Engineering of the eDHFR:Cys variant library
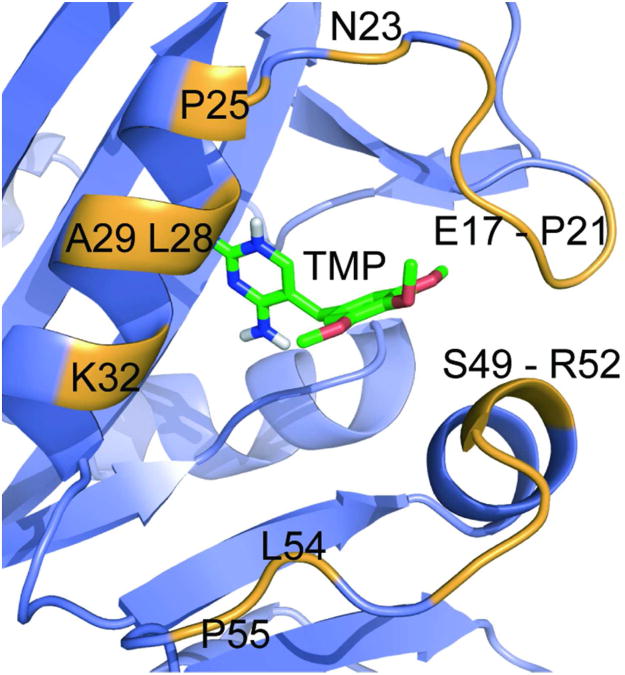
It is well established in the covalent inhibitor literature that the position of the Cys nucleophile strongly affects the reaction rate and labeling efficiency.(22, 26) Thus, we decided to screen a small library of eDHFR variants in search of the eDHFR:Cys variant with maximal reactivity. The eDHFR:Cys variant candidates were selected based on a homology model of the eDHFR/TMP complex. Since no high-resolution crystal structure of eDHFR binding TMP is available, we built an eDHFR/TMP homology model by aligning eDHFR to the highly homologous L. casei DHFR/TMP complex.(30, 31) We have learned from the optimization of covalent TMP-tag that a 10-atom spacer between the 4′-OH group of TMP and the β-carbon of the electrophile would enable rapid proximityinduced reaction.(22) To choose the Cys variants to evaluate, we started from all 39 residues on eDHFR that are within 12 Å (approximately the distance of nine C-C bonds) from the 4′-OH group of TMP. Among these 39 candidates, we selected 16 residues bearing in mind (1) the residue should be on the surface of eDHFR and thus accessible to the electrophile; (2) the residue should orient towards the TMP binding pocket; and (3) mutation of the residue should not significantly impair eDHFR/TMP binding or the stability of eDHFR (Figure 3).(32) The estimated distance between the 4′-OH group of TMP and the 16 amino acid residues are listed in Supplementary Table S1.
Figure 3.
Depiction of the designed eDHFR:Cys mutant library. Cartoon of TMP bound to eDHFR, with all 16 amino acid residues around the binding pocket mutated to Cys highlighted. The eDHFR protein is represented in violet as a ribbon diagram, with the highlighted residues in orange. The TMP is represented as sticks with coloring based on elements. Since no structure has been solved of TMP bound to eDHFR, the model was created by structural alignment of eDHFR with the L. casei DHFR. The graphic was prepared using PyMOL.
Experimentally, we created the eDHFR:Cys mutant library by first fusing the eDHFR gene with a His6-tag in an E. coli expression vector and then mutating 16 amino acids individually using standard site-directed mutagenesis (SDM) technology. We also removed the two naturally occurring cysteines, Cys85 and Cys152, by mutating them to serines to avoid any interference with the cysteine labeling reaction or with protein folding and purification. A variant containing the C85S and C152S mutations, but lacking any other Cys mutation, was used as a control and is referred to as eDHFR-2C.
In vitro characterization of fluorogenic TMP-tags
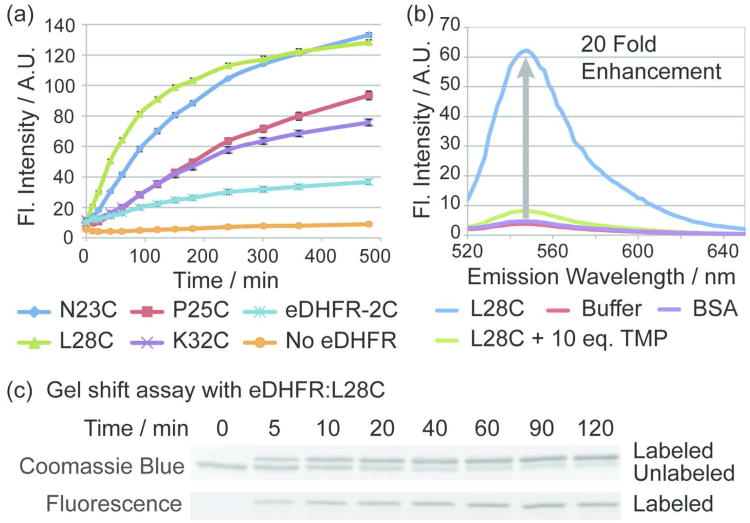
To verify that the designed fluorogenic TMP-tag undergoes efficient and specific quencher cleavage, the TMP-Q-Atto520 was first characterized in vitro using purified eDHFR:Cys variants. By screening the eDHFR:Cys library, we identified eDHFR:L28C as the variant with the fastest reactivity. Then we confirmed its efficiency and specificity using fluorometry and gel shift assays.
The library of 16 different eDHFR:Cys mutants was initially screened by fluorometric measurement to identify the eDHFR:Cys mutant that induces the greatest fluorescence enhancement upon reacting with TMP-Q-Atto520. In brief, 1 μM of each eDHFR:Cys variant was incubated with 1 μM TMP-Q-Atto520 in phosphate buffered saline (PBS) supplemented with 100 μM NADPH and 1 mM reduced glutathione (GSH) at 37 °C. As described for our initial and second-generation covalent TMP-tags, we chose this buffer to mimic the intracellular environment considering (1) NADPH affects eDHFR binding to TMP and its intracellular concentration is close to 100 μM; and (2) GSH represents the pool of various bio-molecules bearing non-specific thiol groups that may potentially compete with the eDHFR:Cys variants for reaction with TMP-Q-Atto520.(21, 22)Figure 4a displays the fluorescence intensity over time for a subset of the eDHFR:Cys variants that induced significant fluorescence enhancement with TMP-Q-Atto520; the full data for all 16 eDHFR:Cys variants can be found in the Supporting Information (Figure S3). Significantly, the TMP-Q-Atto520 showed the greatest fluorescence enhancement with eDHFR:L28C and eDHFR:N23C. As important controls, TMP-Q-Atto520 incubated with buffer only or with the eDHFR-2C control showed little fluorescence enhancement over time, confirming that the BHQ1 quencher can only be cleaved rapidly in the presence of a specific Cys residue engineered on the surface of eDHFR.
Figure 4.
In vitro reactivity of fluorogenic TMP-tag. To demonstrate the specific fluorescence enhancement and efficiency of covalent labeling, TMP-Q-Atto520 was characterized with purified eDHFR:Cys variants in fluorometry and gel-shift assays. a) Screening of the eDHFR:Cys variant library. Purified eDHFR:Cys variants at a concentration of 1 μM were incubated with 1 μM TMP-Q-Atto520 in PBS (pH = 7.40) with 100 μM NADPH and 1 mM glutathione at 37 °C. Fluorescence intensity (Fl. Intensity) was measured at various time points. Four eDHFR:Cys mutants, namely the eDHFR:N23C, P25C, L28C and K32C showed significant fluorescence enhancement. As controls, the eDHFR-2C variant that lacks Cys and the buffer without eDHFR induced little increase in fluorescence intensity. Error bars represent standard deviation. b) The fluorescence emission spectra of TMP-Q-Atto520. Purified eDHFR:L28C was labeled under the same reaction conditions as in (a) for 3 h. The fluorescence spectrum showed 20 fold enhancement compared to the control group in which 1 μM TMP-Q-Atto520 was incubated with the buffer without eDHFR:L28C. By contrast, 1 μM TMP-Q-Atto520 incubated with 1 μM bovine serum albumin (BSA) which does not bind TMP shows no significant fluorescence enhancement compared to control group with buffer. In addition, the specific quencher cleavage is further demonstrated by the competition assay in which 1 μM TMP-Q-Atto520 was incubated with 1 μM eDHFR:L28C and 10 μM TMP. Standard deviation < 5% and error bards are not shown. c) Determination of the rate of covalent labeling. In the same buffer as in (a), 2 μM of eDHFR:L28C was incubated with 10 μM TMP-Q-Atto520 at 37 °C. At various time points, aliquots were quenched by boiling at 95 °C with 6X SDS. The reaction was analyzed by sodium dodecyl sulfate (SDS)-PAGE followed by fluorescence gel scanning and Coomassie staining, and it was determined that 50% labeling occurs in approximately 10 minutes.
The best variant eDHFR:L28C was further evaluated with TMP-Q-Atto520 for labeling efficiency and specificity. We first measured fluorescence excitation (Supplementary Figure S4) and emission spectra (Figure 4b) under the same labeling conditions as described above. Fluorescence enhancement (EF) was calculated as EF = F/F0−1, in which F is the fluorescence intensity of TMP-Q-Atto520 labeling the eDHFR:L28C; F0 is the fluorescence intensity of the control group with buffer and GSH while lacking eDHFR protein. As presented in Figure 4b, 1 μM eDHFR:L28C incubated with 1 μM TMP-Q-Atto520 for 3 h at 37 °C produced 20 fold fluorescence enhancement. Importantly, we also performed a TMP competition assay in which TMP-Q-Atto520 was incubated with eDHFR:L28C in the presence of various concentrations of TMP. As reported in Figure 4b as well as the Supporting Information (Figure S5), 1 – 10 equivalents of TMP could efficiently compete off labeling of eDHFR:L28C by TMP-Q-Atto520, further verifying that the mechanism of the fluorogenic TMP-tag is proximity-induced labeling of eDHFR:L28C.
The covalent and site-specific labeling of eDHFR:L28C by fluorogenic TMP-tag was further validated by mass spectroscopy (MS) analysis. Primarily, the molecular weight of eDHFR:L28C labeled with fluorogenic TMP-tag was measured using matrix-assisted laser desorption/ionization (MALDI) MS in comparison with unlabeled eDHFR:L28C, showing an increase in molecular weight as anticipated for covalent conjugation with the fluorogenic TMP-tag. Then the labeled eDHFR:L28C was digested by chymotrypsin followed by liquid chromatography-tandem MS (LC-MS/MS) analysis to identify the amino acid residue being labeled. The fragmentation pattern revealed by MS/MS was consistent with L28C labeling (Supplementary Figure S6).
Next we measured the kinetics of the labeling reaction by gel-shift assay. Specifically, the in vitro eDHFR:L28C labeling reaction with TMP-Q-Atto520 was quenched at various time points and analyzed with SDS-PAGE. With Coomassie staining, labeling of eDHFR:L28C produced a band-shift on the gel presumably due to the increase in molecular weight of the labeled protein, allowing the progress of the reaction to be monitored by densitometry analysis. Significantly, both in-gel fluorescence and gel-shift assay demonstrated the accumulation of covalently labeled eDHFR:L28C over time (Figure 4c). The half time of the labeling reaction (t1/2) was estimated to be ∼ 10 min for TMP-Q-Atto520 labeling eDHFR:L28C. And the rate constant for the labeling reaction has been estimated to be 53 M−1s−1 assuming a secondorder reaction mechanism (Supplementary Figure S7). Taken together, the in vitro assays confirmed that TMP-Q-Atto520 rapidly produced a highly specific, covalent label with significant fluorescence enhancement, suggesting it would be robust enough to label proteins in living cells.
Importantly, while this initial report focuses on TMP-Q-Atto520, the fluorogenic TMP-tag is designed to be modular and should allow for other organic fluorophores to be readily introduced. Thus, to verify the modularity of the fluorogenic design, we also synthesized a TMP-Q-fluorescein tag that is identical to TMP-Q-Atto520, just with a different fluorophore. We characterized the TMP-Q-fluorescein in vitro following all of the assays described above. As expected, the TMP-Q-fluorescein also demonstrated specific and rapid labeling, in this case with > 50 fold fluorescence enhancement – the background fluorescence is significantly lower than for TMP-Q-Atto520 (Supplementary Figure S8, S9). Interestingly, the reactivity of TMP-Q-fluorescein with the eDHFR:Cys variants was distinct compared to TMP-Q-Atto520. In brief, the TMP-Q-fluorescein labeled eDHFR:P25C faster than all other mutants, followed by eDHFR:L28C, and the reaction half time was estimated to be ∼ 50 min with eDHFR:P25C and ∼ 150 min with eDHFR:L28C – slower than for TMP-Q-Atto520 (see Supplementary Figure S10). One explanation for this difference in reactivity is that the proximity-induced reaction is affected by electrostatic interactions between the fluorophore and eDHFR — fluorescein is negatively charged while Atto520 is positively charged at physiological pH. Notably, the TMP-Q-fluorescein also enabled nuclear and plasma membrane-localized eDHFR to be labeled in live HEK 293T cells with high efficiency and specificity (Supplementary Figure S11), although its applications in live cell imaging of diffused cytosolic proteins were limited by the modest cell permeability of the negatively charged fluorescein. Together these results confirm that our fluorogenic design is modular and can be readily adapted to new fluorophores with diverse chemical properties.
Live cell imaging
Encouraged by the efficient and rapid labeling reaction between eDHFR:L28C and TMP-Q-Atto520 with strong fluorescence enhancement, we next evaluated the potential of fluorogenic TMP-tag for live cell imaging with high S/B. We started by labeling a nuclear localized target protein and showed the fluorogenic TMP-Q-Atto520 specifically stains the target protein in live cells with significantly lower background compared to the non-fluorogenic TMP-Atto520. Then we confirmed the robustness of the fluorogenic TMP-tag by successfully labeling cytosolic proteins that are more diffuse and dynamic, characterized by both microscopy and in-gel fluorescence analysis.
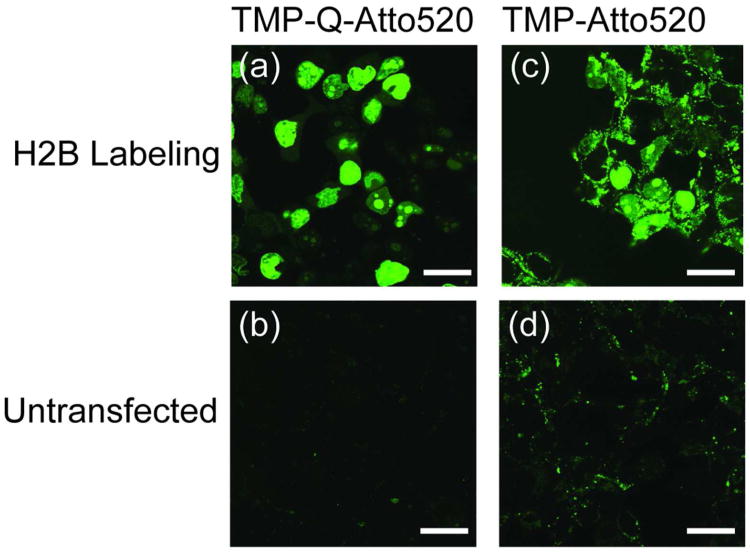
First we evaluated the efficiency and selectivity of TMP-Q-Atto520 for labeling nuclear localized human histone H2B in HEK 293T cells. Specifically, HEK 293T cells were transiently transfected with plasmid encoding H2B-eDHFR:L28C and incubated with 5 μM TMP-Q-Atto520 in tissue culture media. To compare the fluorogenic TMP-tag with non-fluorogenic TMP-tag, we transfected another group of cells with H2B-eDHFR (wild type) and labeled them with 5 μM TMP-Atto520. As controls, untransfected HEK 293T cells were stained with 5 μM TMP-Q-Atto520 or 5 μM TMP-Atto520, respectively (Figure 5). For comparison, all images in Figure 5 were taken under identical microscope setting including the objective, laser intensity, pinhole size, and gain value of the photon multiplier tube (PMT). Remarkably, the TMP-Q-Atto520 efficiently labeled the nucleus of live cells with no visible background staining. By contrast, the non-fluorogenic TMP-Atto520 displayed high background fluorescence from non-specific staining and/or unbound fluorophores. In the control groups with untransfected cells, the TMP-Atto520 also showed much stronger non-specific staining than the fluorogenic TMP-Q-Atto520.
Figure 5.
Live cell imaging of cell nucleus with fluorogenic TMP-tag in comparison with non-fluorogenic TMP-tag. a) HEK 293T cells were transiently transfected with plasmid encoding histone 2B (H2B)-eDHFR:L28C and labeled with 5 μM TMP-Q-Atto520 for 3 h. Live cell imaging was achieved using confocal microscope, displaying the specific labeling of cell nucleus. b) As control, untransfected cells were treated with TMP-Q-Atto520 under the same condition as (a), showing very weak background fluorescence. c, d) The same imaging experiments were carried out with the first generation, noncovalent TMP-tag. Significant background staining was observed with both H2B-eDHFR transfected and untransfected cells. For comparison, all four images were obtained using the same microscope setup with excitation at 488 nm. Scale bars are 25 μm.
In addition, we also evaluated the robustness of the fluorogenic TMP-tag by carrying out live cell imaging under various conditions. Significantly, labeling H2B-eDHFR:L28C with low concentration (1 μM) of TMP-Q-Atto520 or short staining time (1 h) still enabled live cell imaging with high S/B (Supplementary Figure S12). The labeling efficiency in live HEK 293T cells has been quantified using western blot: 38% of intracellular eDHFR:L28C was covalently labeled within 1 h; 3 h staining with 10 μM TMP-Q-Atto520 resulted in 67% labeling yield (Supplementary Figure S13). Based on these results we recommend 5 –10 μM of TMP-Q-Atto520 staining for 2 – 3 h to provide maximal S/B for live cell imaging.
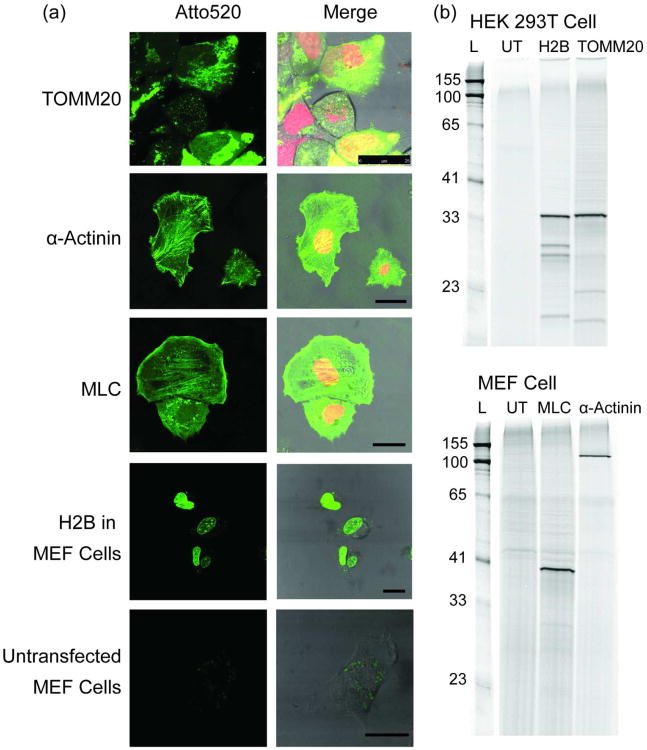
After demonstrating the advantage of the fluorogenic TMP-Q-Atto520 over non-fluorogenic TMP-Atto520, we further assessed the robustness of the fluorogenic TMP-tag by labeling a series of different proteins in various cell lines. The target proteins we tested included the translocase of outer mitochondrial membrane 20 (TOMM20); myosin light chain (MLC); and α-actinin. All of these target proteins are less abundant and more dynamic than H2B, making them more challenging targets for live cell imaging. To label the target proteins with fluorogenic TMP-tag, HEK 293T cells were transiently transfected with plasmids encoding TOMM20-eDHFR:L28C and MEF cells were electroporated to introduce plasmids encoding MLC-eDHFR:L28C and α-actinin-eDHFR:L28C. All cells were also transfected with a control vector encoding H2B-mCherry as the transfection indicator. As presented in Figure 6a, TMP-Q-Atto520 labeled all target proteins with high S/B, showing the desired sub-cellular localizations: TOMM20 at mitochondria that distributed around the nucleus; MLC in stress fibers as well as lamilipodia; α-actinin (an actin crosslinker) developing the ‘dash line’ pattern along the stress fibers and also localized to the focal adhesions.(33-36) In all experiments we observed no evidence of cytotoxicity, as the cells labeled with TMP-Q-Atto520 showed no loss of viability or any other phenotypic variations compared with mock treated control cells (Supplementary figure S14). The correct sub-cellular localization of target proteins also indicated no loss or alternation in protein folding and function.
Figure 6.
Live cell imaging of cytosolic proteins with TMP-Q-Atto520. Three cytosolic proteins, TOMM20, α-actinin, MLC, in addition to H2B were successfully imaged in two distinct cell lines. a) Live cell imaging using TMP-Q-Atto520. HEK 293T cells (for TOMM20) or mouse embryonic fibroblast (MEF) cells (for MLC, α-actinin, and H2B) transiently co-transfected with vectors encoding target protein-eDHFR:L28C and H2B-mCherry (for TOMM20, MLC and α-actinin labeling) fusion proteins, respectively, were incubated with 10 μM TMP-Q-Atto520 in media for 3 h at 37 °C, washed twice with media, and imaged using confocal and differential interference contrast (DIC) microscopy. Untransfected MEF cells were treated under the same condition and imaged using the same microscope setup as for H2B in MEF cells. Left column shows fluorescence imaging from the green channel (488 nm excitation); right column shows merges images from the green, red (594 nm excitation), and DIC channels. Scale bars are 25 μm. b) In-gel fluorescence analysis to confirm the labeling specificity. The cells transfected with corresponding target protein-eDHFR:L28C vectors were harvested after 3 h incubation with 10 μM TMP-Q-Atto520, lysed, and analyzed by SDS-PAGE and in-gel fluorescence scanning with an excitation laser at 488 nm. As controls, untransfected (UT) cells were stained with TMP-Q-Atto520 under the same condition and analyzed in parallel. These results show the target versatility of the fluorogenic TMP-tag for live cell protein labeling.
Finally, the specificity of TMP-Q-Atto520 labeling in living cells was examined by SDS-PAGE and in-gel fluorescence scanning. The HEK 293T cells and MEF cells were transfected and labeled with TMP-Q-Atto520 following the same procedures as for live cell imaging. As negative controls, untransfected HEK 293T cells and MEF cells were stained with TMP-Q-Atto520 as well. After cell lysis, the supernatants of the lysates were loaded onto SDS-PAGE and the gels were scanned for in-gel fluorescence. As presented in Figure 6b, in all lanes only one major band correlating to the expected fusion protein was observed: 33 kDa H2B-eDHFR:L28C; 34 kDa TOMM20-eDHFR:L28C; 37 kDa MLC-eDHFR:L28C; 121 kDa α-actinin-eDHFR:L28C. The absence of any visible band for both untransfected HEK 293T cells and MEF cells confirms the high specificity of TMP-Q-Atto520. Interestingly, there are a few minor bands with lower molecular weight in the lysate of the cells transfected with H2B-eDHFR:L28C and TOMM20-eDHFR:L28C. Because these lower molecular weight bands are absent in other supernatants, we interpret these bands as degradation products of the fusion protein. These results demonstrate that TMP-Q-Atto520 efficiently forms a covalent bond with intracellular eDHFR:L28C fusion proteins with high specificity, verifying the utility of the fluorogenic TMP-tag as a robust cellular reagent for live cell imaging.
Discussion
Together these results establish that TMP-Q-Atto520 is a viable fluorogenic chemical tag for live cell imaging with high S/B. The in vitro labeling reaction is rapid (t1/2 ∼ 10 min) and efficient even in the presence of high concentrations of thiol species. The proximity-induced quencher displacement yields a 20 fold fluorescence enhancement for TMP-Q-Atto520, which means 20 fold increase in S/B compared to the non-fluorogenic TMP-tag. Perhaps most significantly, TMP-Q-Atto520 is sufficiently reactive and selective to enable live cell imaging with high S/B, labeling not only nuclear localized H2B, but also diffuse cytosolic proteins involved in cell motility.
We envision that the fluorogenic TMP-tag will be a valuable tool for SM imaging. TMP- tag is an attractive chemical tag because eDHFR is a small (18 kD), monomeric protein and TMP analogs are readily cell-permeable, straightforward to synthesize, and show no apparent cross-reactivity or toxicity in mammalian cells. Working with collaborators we have begun to tackle sophisticated biophysical experiments in live cells, including SM analysis of spliceosome assembly in yeast cell extracts and the molecular mechanism of mechanical sensing in live mammalian cells.(13, 37) These SM studies will significantly benefit from the improvement in S/B brought by the fluorogenic TMP-tag.
The fluorogenic TMP-tag features a modular design that can be readily adapted to new fluorophores spanning the whole visible-infra red spectral region. To date, reported fluorogenic probes based on solvatochromes, molecular rotors and PeT mostly hinge on the special charge distribution or isomerization of the fluorogen.(38) As a result, it is challenging to generalize the fluorogenic design to new fluorophores at different wavelengths, especially those with high photon outputs and the desired photo-physical properties for high-resolution imaging. In contrast, the cleavable quencher strategy is more generalizable. Fluorogenic chemical tags based on cleavable quenchers have been reported recently for SNAP-tag and a β-lactamase tag, although these were mainly used to label cell surface or nuclear localized proteins.(39-42) With a fluorogenic TMP-tag based on a cleavable quencher that works inside cells, we should be in place to build a series of fluorogenic TMP-tags using different fluorophores for multi-color SM imaging.
The fluorogenic design minimizes non-specific background staining of cellular compartments often observed for the highly charged, high photon-output fluorophores. On its own TMP-Atto520, like many high photon output fluorophores, has a delocalized positive charge on the fluorophore, rendering the tag readily cell permeable but prone to aggregation and non-specific staining of mitochondria. The fluorogenic design suppresses the background fluorescence from unbound and non-specifically bound TMP-Atto520, making TMP-Q-Atto520 a viable reagent for live cell imaging. Going forward, we are exploring the potential of this fluorogenic design to make other high photon output fluorophores that also suffer from non-specific staining of cellular compartments, such as the cyanine and oxazine-based fluorophores, viable reagents for SM imaging in live cells.
Furthermore, the elegance of the proximity-induced reactivity strategy is that it can be easily extended to converting other high affinity ligand/receptor pairs into new chemical tags that are orthogonal to the fluorogenic TMP-tag. For the fluorogenic TMP-tag, the remarkable selectivity is guaranteed by high affinity enzyme/inhibitor recognition; the efficiency comes from the proximity-facilitated SN2 reaction between the engineered protein Cys nucleophile and mild ligand electrophile (tosylate). Using similar approaches, the vast pool of drug/receptor pairs could be potentially utilized to build a toolbox for multi-color SM imaging in live cells, to provide insight into the molecular mechanism of complex biological pathways.
Finally, the implementation of fluorogenic TMP-tag for live cell imaging further validates the proximityinduced reaction as a viable strategy to develop chemical tools for probing and manipulating live cells. We have previously engineered the covalent TMP-tag based on proximity-induced Michael addition, enabling high resolution live cell imaging of a variety of target proteins with modular organic fluorophores. Now we introduce the SN2 reaction – another type of classic reaction in organic synthesis – as a modular tool for site-specific modification of various intracellular biomolecules. Together this work shows that organic chemistry can be added to the arsenal of tools available for synthetic biology and cell engineering, as it has thrived in the field of protein and nucleic acid engineering.
Conclusion
Here we successfully developed a fluorogenic TMP-tag that is sufficiently reactive and selective to enable live cell imaging of intracellular proteins with high S/B. Remarkably, TMP-Q-Atto520 was shown to be readily cell permeable and well-behaved in live cells, such that not only localized but also diffuse cytosolic proteins can be labeled via a straightforward staining procedure. The highly modular design of our fluorogenic TMP-tag should allow it to be readily generalized to new chemical tags with different fluorophores and orthogonal ligand/receptor pairs. Using this strategy we envision constructing a toolbox of several orthogonal chemical tags for multi-color SM imaging in live cells, which will greatly impact the field of biophysics and cell biology. Just as chemical methods for selectively labeling proteins in the test tube deeply impacted in vitro biophysics in the last century, chemical tag technologies have the potential to enable organic chemistry to provide the breakthrough needed to meet this century's challenge of understanding multi-component biological pathways in the living cell.
Methods
Methods for chemical synthesis, site-directed mutagenesis, protein purification, in vitro characterization and live cell imaging are provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH 5RC1GM091804). We would like to thank M.P. Sheetz for providing the MEF cell culture and advice on designing the fluorogenic TMP-tag. We thank K. Yeager for support for confocal microscopy. We thank T.Y. Wang and Z. Chen for their helpful advice on the experiments and manuscript. We thank M.A. Gawinowicz in Protein Core Facility of Columbia University for carrying out MS and LC-MS/MS analysis. Finally, we are indebted to S.S. Gallagher and R. Wombacher for their contribution to this project at the early stage.
Footnotes
Supporting Information: This material is free of charge via the Internet: http://pubs.acs.org
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 4.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 5.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 6.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Jing CR, Cornish VW. Chemical Tags for Labeling Proteins Inside Living Cells. Acc Chem Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 9.Calloway NT, Choob M, Sanz A, Sheetz MP, Miller LW, Cornish VW. Optimized fluorescent trimethoprim derivatives for in vivo protein labeling. Chembiochem. 2007;8:767–774. doi: 10.1002/cbic.200600414. [DOI] [PubMed] [Google Scholar]
- 10.Roth B, Aig E, Rauckman BS, Strelitz JZ, Phillips AP, Ferone R, Bushby SR, Sigel CW. 2,4-Diamino-5-benzylpyrimidines and analogues as antibacterial agents. 5 3′,5′-Dimethoxy-4′-substituted-benzyl analogues of trimethoprim. J Med Chem. 1981;24:933–941. doi: 10.1021/jm00140a005. [DOI] [PubMed] [Google Scholar]
- 11.Baccanari DP, Daluge S, King RW. Inhibition of Dihydrofolate-Reductase - Effect of Reduced Nicotinamide Adenine-Dinucleotide Phosphate on the Selectivity and Affinity of Diaminobenzylpyrimidines. Biochemistry. 1982;21:5068–5075. doi: 10.1021/bi00263a034. [DOI] [PubMed] [Google Scholar]
- 12.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Live-cell super-resolution imaging with trimethoprim conjugates. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 13.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reymond JL, Fluxa VS, Maillard N. Enzyme assays. Chem Commun. 2009:34–46. doi: 10.1039/b813732c. [DOI] [PubMed] [Google Scholar]
- 15.Shestopalov IA, Chen JK. Spatiotemporal Control of Embryonic Gene Expression Using Caged Morpholinos. Method Cell Biol. 2011;104:151–172. doi: 10.1016/B978-0-12-374814-0.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsien RY. Calcium as a cellular regulator. Oxford University Press; New York: 1999. [Google Scholar]
- 17.Loving GS, Sainlos M, Imperiali B. Monitoring protein interactions and dynamics with solvatochromic fluorophores. Trends Biotechnol. 2010;28:73–83. doi: 10.1016/j.tibtech.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haidekker MA, Lichlyter D, Ben Johny M, Grimes CA. Probing polymerization dynamics with fluorescent molecular rotors and magnetoelastic sensors. Sens Lett. 2006;4:257–261. [Google Scholar]
- 19.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 20.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 21.Gallagher SS, Sable JE, Sheetz MP, Cornish VW. An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem Biol. 2009;4:547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ZX, Jing CR, Gallagher SS, Sheetz MP, Cornish VW. Second-Generation Covalent TMP-Tag for Live Cell Imaging. J Am Chem Soc. 2012;134:13692–13699. doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang XW. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Linde S, Endesfelder U, Mukherjee A, Schuttpelz M, Wiebusch G, Wolter S, Heilemann M, Sauer M. Multicolor photoswitching microscopy for subdiffraction-resolution fluorescence imaging. Photoch Photobio Sci. 2009;8:465–469. doi: 10.1039/b822533h. [DOI] [PubMed] [Google Scholar]
- 25.Marras SAE, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitsky K, Boersma MD, Ciolli CJ, Belshaw PJ. Exo-mechanism proximity-accelerated alkylations: Investigations of linkers, electrophiles and surface mutations in engineered cyclophilin-cyclosporin systems. Chembiochem. 2005;6:890–899. doi: 10.1002/cbic.200400383. [DOI] [PubMed] [Google Scholar]
- 27.Tsukiji S, Wang HX, Miyagawa M, Tamura T, Takaoka Y, Hamachi I. Quenched Ligand-Directed Tosylate Reagents for One-Step Construction of Turn-On Fluorescent Biosensors. J Am Chem Soc. 2009;131:9046–9054. doi: 10.1021/ja902486c. [DOI] [PubMed] [Google Scholar]
- 28.Tsukiji S, Miyagawa M, Takaoka Y, Tamura T, Hamachi I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nat Chem Biol. 2009;5:341–343. doi: 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- 29.Cook RM, Lyttle M, Dick D. In: Dark quenchers for donor-acceptor energy transfer. Office, U. S. P. a. T., editor. Biosearch Technologies, Inc; United States: 2000. [Google Scholar]
- 30.Polshakov VI, Smirnov EG, Birdsall B, Kelly G, Feeney J. Letter to the Editor: NMR-based solution structure of the complex of Lactobacillus casei dihydrofolate reductase with trimethoprim and NADPH. J Biomol NMR. 2002;24:67–70. doi: 10.1023/a:1020659713373. [DOI] [PubMed] [Google Scholar]
- 31.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: Crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 32.Matthews DA, Bolin JT, Burridge JM, Filman DJ, Volz KW, Kaufman BT, Beddell CR, Champness JN, Stammers DK, Kraut J. Refined crystal structures of Escherichia coli and chicken liver dihydrofolate reductase containing bound trimethoprim. J Biol Chem. 1985;260:381–391. [PubMed] [Google Scholar]
- 33.von Wichert G, Haimovich B, Feng GS, Sheetz MP. Force-dependent integrin-cytoskeleton linkage formation requires downregulation of focal complex dynamics by Shp2. EMBO J. 2003;22:5023–5035. doi: 10.1093/emboj/cdg492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 35.Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grosheva I, Vittitow JL, Goichberg P, Gabelt BT, Kaufman PL, Borras T, Geiger B, Bershadsky AD. Caldesmon effects on the actin cytoskeleton and cell adhesion in cultured HTM cells. Exp Eye Res. 2006;82:945–958. doi: 10.1016/j.exer.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Cai Y, Rossier O, Gauthier NC, Biais N, Fardin MA, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukinavicius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Correa IR, Jr, Luo ZG, Schultz C, Lemke EA, Heppenstall P, Eggeling C, Manley S, Johnsson K. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem. 2013;5:132–139. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- 39.Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng LX, Whitney M, Roemer K, Tsien RY. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 40.Komatsu T, Johnsson K, Okuno H, Bito H, Inoue T, Nagano T, Urano Y. RealTime Measurements of Protein Dynamics Using Fluorescence Activation-Coupled Protein Labeling Method. J Am Chem Soc. 2011;133:6745–6751. doi: 10.1021/ja200225m. [DOI] [PubMed] [Google Scholar]
- 41.Sun XL, Zhang AH, Baker B, Sun L, Howard A, Buswell J, Maurel D, Masharina A, Johnsson K, Noren CJ, Xu MQ, Correa IR. Development of SNAP-Tag Fluorogenic Probes for Wash-Free Fluorescence Imaging. Chembiochem. 2011;12:2217–2226. doi: 10.1002/cbic.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizukami S, Watanabe S, Akimoto Y, Kikuchi K. No-wash protein labeling with designed fluorogenic probes and application to real-time pulse-chase analysis. J Am Chem Soc. 2012;134:1623–1629. doi: 10.1021/ja208290f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.