Abstract
Due to the vital roles proteins play in life, much effort has been invested in their mimicry by synthetic agents. One approach to this goal is to design unnatural backbone oligomers (“foldamers”) that fold like natural peptides. Despite success in secondary structure mimicry by such species, protein-like tertiary folds remain elusive. A fundamental challenge underlying this task is the design of a sequence of side chains that will specify a complex tertiary folding pattern on an unnatural backbone. We report here a sequence-based approach to convert a natural protein with a compact tertiary fold to an analogue with a backbone composed of ∼20% unnatural building blocks but similar folding behavior as the parent protein.
The diversity of proteins is vast, but their behavior is based on a simple dogma: biological function is determined by a three-dimensional fold specified, in turn, by a sequence of amino acids.1 Recent years have seen the development of unnatural oligomers with protein-like folds and functions (termed “foldamers”).2 From a fundamental perspective, such work demonstrates the degree to which natural biopolymers are unusual in their ability to manifest diverse folds from a common oligomeric backbone. From a practical standpoint, unnatural species that mimic proteins represent starting points toward bioactive agents capable of recognizing protein surfaces involved in protein–protein binding interactions,2c-e a challenging target class for therapeutic development.3
A central question in the mimicry of proteins by foldamers is how to design a sequence of building blocks that will fold like a particular α-peptide. Strategies have been reported for mimicry of secondary structure by unnatural backbones,2 but the task becomes more daunting as the target fold increases in complexity. These more complex folded structures are important targets, since many biological functions involve tertiary or quaternary folds. Reports of unnatural oligomers with folded complexity beyond secondary structure include helix-bundle assemblies,4 helix-turn-helix motifs,5 and a non-regular folding pattern from a disulfide-cyclized species.6 There remains an unmet need for a general design strategy for mimicry of a typical protein tertiary fold (helix, loop, sheet, and turn secondary structures in a single chain lacking disulfides) by an oligomer with an unnatural backbone.
Our goal is to develop a general approach, based on natural sequence information, for the mimicry of protein tertiary folding by unnatural oligomers. Early foldamer research focused on homogeneous backbones made up entirely of unnatural building blocks, but recent work has shown the utility of blending natural and unnatural residues to create heterogeneous backbones.7 Folded proteins can tolerate a variety of backbone alterations;8 however, such modifications are typically made in isolation. Underlying our design strategy is the hypothesis that any protein can be thought of as an oligomer with two mutually orthogonal sequences: (1) the canonical sequence of amino acid sidechain functional groups and (2) a separate sequence of backbone elements that display those side-chain functional groups. In nature, “backbone sequence” (2) is an invariant l-α-peptide, but synthetic oligomers are not subject to this limitation. Here, we systematically alter backbone and side-chain sequence in a bacterial protein with a defined tertiary folding pattern to generate an analogue with ∼20% unnatural backbone but similar folding behavior as the parent protein.
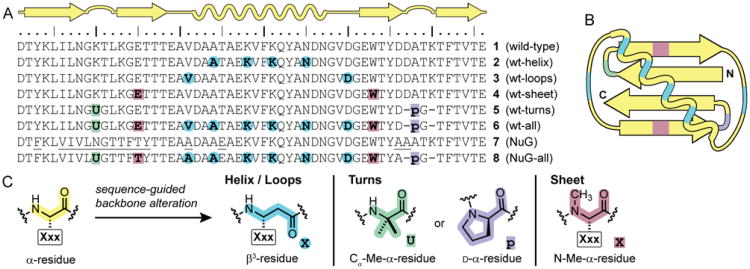
We selected the B1 domain of Streptococcal protein G (GB1, 1) as a model system. The native folded state of GB1 contains all the common types of secondary structure in a well-defined tertiary fold devoid of disulfide bridges (Figure 1A, B).9 Although simple when compared to the array of known protein folds, GB1 is at the frontier of complexity in folding patterns demonstrated by unnatural backbone oligomers. From the wild-type sequence (1), we designed several variants to explore the impact of backbone alteration on folded structure and stability (Figure 1). GB1 analogues 2-6 incorporate different patterns of unnatural backbone substitutions into the wild-type side-chain sequence. All proteins were synthesized by Fmoc solid-phase methods, purified by a two-stage chromatographic protocol, and their identities confirmed by mass spectrometry. Alterations to the backbone in these proteins include carbon insertion (β3-residues), inversion of Cα stereochemistry (d-α-residues), and alkylation (Cα-methyl or N-methyl α-residues). These readily accessible building blocks have all been widely explored in short peptides, but most have not been examined in folded proteins. Our aim here is to demonstrate that they can be used in combination as part of a systematic strategy for protein backbone modification.
Figure 1.
(A) Sequence and secondary structure map for wild-type GB1 (1), backbone modified GB1 analogues 2-6, engineered GB1 mutant “NuG” (7), and backbone modified NuG analogue 8. Underlined residues in NuG (7) indicate side-chain mutations relative to wild-type GB1 (1). (B) Tertiary folding topology of GB1 colored by backbone composition of analogues 6 and 8. (C) Key to residue types used in sequence-guided backbone alteration. For β3-residues and N-Me-α-residues, the side chain “Xxx” is the same as that of the replaced natural amino acid “X”.
Prior work has shown that α/β-peptides derived from helix-forming natural sequences can adopt native-like folded structures that retain biological function.4b,10 We reasoned that this strategy for secondary structure mimicry would also be successful in a context, such as GB1, where the helix is involved in key long-range contacts that define a tertiary folding pattern. In order to test this hypothesis, we prepared protein 2. In analogue 2, sequence guided α→β3 residue replacements are applied to a subset of α-residues in the GB1 helix. Each replaced α-residue is substituted with a β3-residue bearing the side chain of the parent amino acid. The α→β3 substitutions were applied in an ααβαααβ pattern that is based on the heptad repeat of the α-helix and intended to place the additional methylene units at positions in the helix not directly involved in hydrophobic core packing interactions with the sheet in the tertiary fold.
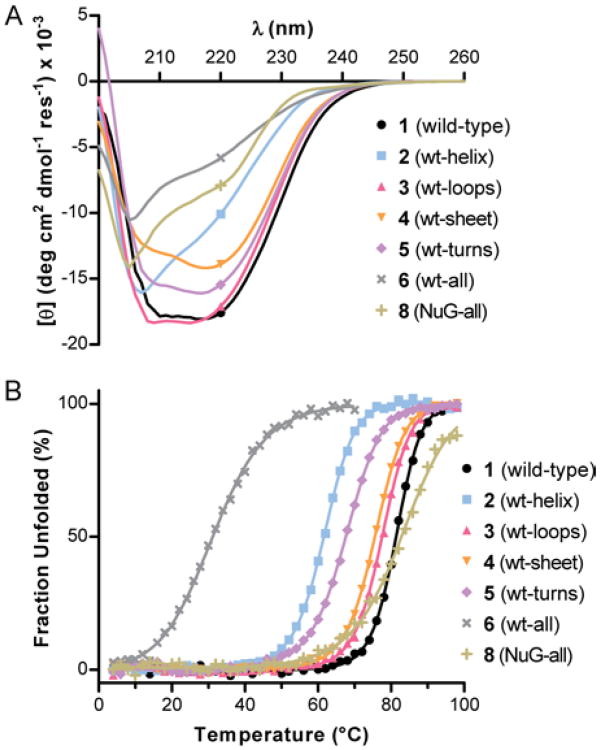
We examined the folding behavior of protein 2 in solution by circular dichroism (CD) spectroscopy. Compared to wild-type GB1, the CD signature of 2 is different in shape and magnitude (Figure 2A); however, the differences are consistent with prior data for α/β-peptide helix bundles.4b,10 CD thermal denaturation experiments monitored at 220 nm revealed a sigmoidal unfolding transition for 2 with cooperativity similar to that of wild-type 1 (Figure 2B). The midpoint of the thermal transition (Tm) was ∼20 °C lower for 2 compared to 1, corresponding to a loss of folding free energy of ∼0.8 kcal/mol per α→β3 residue substitution (Table 1). This destabilization is consistent with prior qualitative observations on α→β3 replacements in helix bundles4b,10 and provides a quantitative measure of the thermodynamic consequence of CH2 insertion in a natural helix backbone.
Figure 2.
(A) CD scans at 25 °C and (B) CD thermal unfolding transitions monitored at 220 nm. Lines in the thermal denaturation data are fits to a two-state unfolding equilibrium. Each sample is 40 μM protein in 20 mM phosphate pH 7.
Table 1.
Folding thermodynamics of wild-type GB1 and backbone-modified analogues.
| Protein | 2° Structure Modified | Tm (°C)a | ΔTm (°C)b | ΔΔGfold (kcal mol-1)c |
|---|---|---|---|---|
| 1 | none | 81.4 ± 0.1 | − | − |
| 2 | Helix | 61.6 ± 0.1 | −19.8 | +3.2 ± 0.1 |
| 3 | Loops | 77.6 ± 0.1 | −3.8 | +0.6 ± 0.1 |
| 4 | Sheet | 75.6 ± 0.1 | −5.8 | +0.9 ± 0.1 |
| 5 | Turns | 67.6 ± 0.1 | −13.8 | +2.2 ± 0.1 |
| 6 | All | ∼31d | −50 | +8 |
| 8 | All | ∼82e | ∼0 | ∼0 |
Midpoint of the thermal transition observed by CD at 220 nm for a 40 μM protein sample in 20 mM phosphate pH 7; estimated errors are uncertainty from the fit.
Change in Tm vs. wild-type (1).
Change in folding free energy vs. 1.
A complete folded baseline was not observed.
A complete unfolded baseline was not observed.
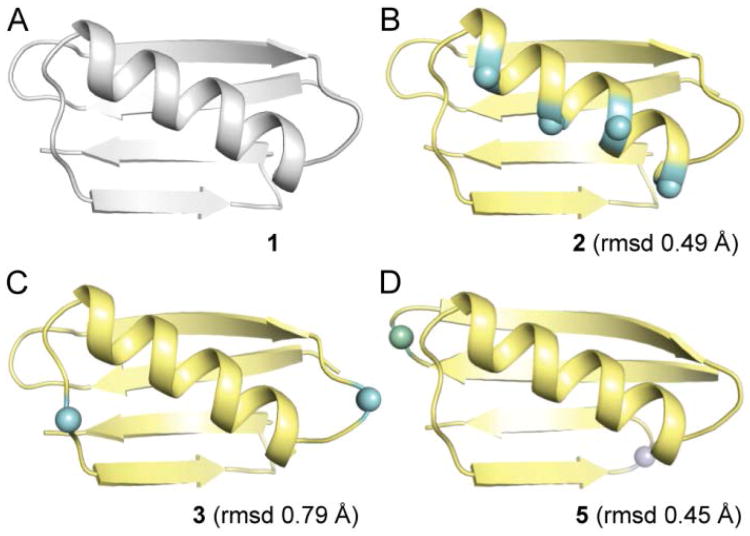
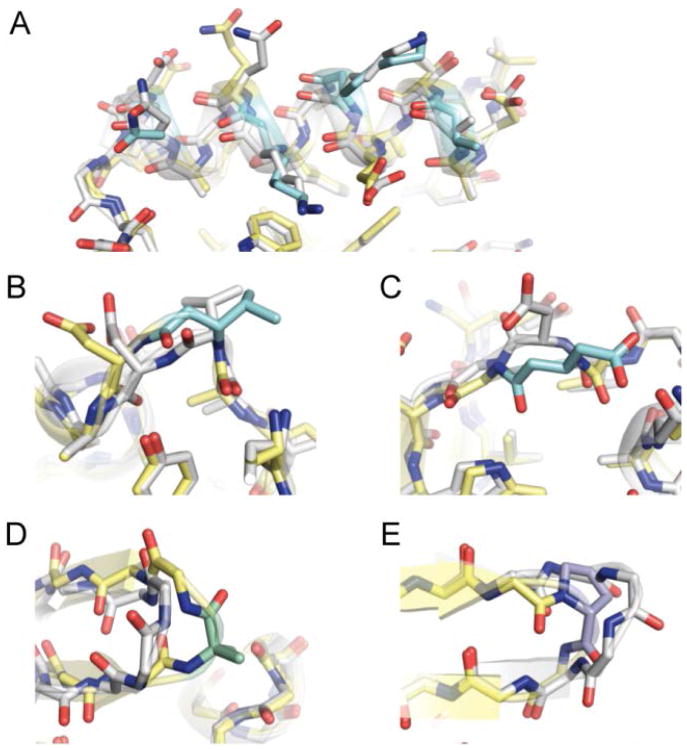
We obtained single crystals of protein 2 by hanging drop vapor diffusion, X-ray diffraction data were collected, and the structure was refined to 2.0 Å resolution (Table S1). The tertiary folding pattern of helix-modified GB1 analogue 2 is virtually identical to wild-type GB1 1 (Figure 3A,B). As predicted, the β3-residues in 2 align on the face of the helix opposite the hydrophobic core. The modified helix maintains the hydrogen-bonding pattern and side-chain contacts of corresponding region in the parent protein (Figure 4A). Particularly noteworthy are the contacts involving sequence position 31. In wild-type GB1 (1), the side chain of Lys31 is packed against core residue Trp43 in the sheet; a similar contact is observed for β3-Lys31 residue in protein 2 (Figure S1). These observations show how backbone alterations that retain side chain information (such as α→β3 replacement) create the possibility to maintain interactions that contribute to folding behavior of a prototype natural sequence on a modified backbone.
Figure 3.
Crystal structures of wild-type GB1 (1, PDB 2QMT)9b and three analogues with modified backbone in the helix (2), loops (3), or turns (5). Root mean square deviation (rmsd) values are calculated for overlay of Cα atoms between 1 and the indicated analogue. Unnatural residues are shown as spheres; the coloring scheme for proteins 2, 3, and 5 matches Figure 1C.
Figure 4.
Crystal structure overlays of wild-type GB1 (1, white carbons) with the indicated segment of backbone-modified analogues 2, 3, or 5 (carbon coloring scheme from Figure 1C): (A) the helix from 2, (B) the N-terminal loop from 3, (C) the C-terminal loop from 3, (D) the N-terminal turn from 5, and (E) the C-terminal turn from 5.
Our prior work has shown that the two surface-exposed loops in GB1 can be replaced by oligomers based on poly(ethylene glycol) (PEG).11 Although the PEG-based GB1 chimeras retained the fold of the native sequence, they were significantly destabilized. In an effort to devise a more energetically neutral loop modification strategy, we applied sequence-guided 1:1 α→β3 residue replacement, as described above for the GB1 helix. One residue in each loop of GB1 (Val21 and Asp40) was replaced with the corresponding β3-residue to generate variant 3 (Figure 1). Loop-modified GB1 analogue 3 showed a nearly identical CD signature as native 1 (Figure 2A), and its Tm suggested a folded stability within 0.6 kcal/mol of wild-type (Figure 2B, Table 1).
We obtained diffraction quality crystals of 3 and solved its structure to 1.95 Å resolution (Table S1). The fold of 3 (Figure 3C) is very similar to that of GB1. Main-chain hydrogen bonds and side-chain interactions near the N-terminal loop are unperturbed (Figure 4B). By contrast, differences are apparent between the structures in the vicinity of the C-terminal loop (Figure 4C). In wild-type GB1 (1), Asp40 is involved in a polar contact with the side chain of Glu56. In variant 3, this contact is lost but new polar contacts are gained (Figure S2). Comparison of the interactions involving Asp40 in 1 vs. β3-Asp40 in 3 show that natural side chain sequences on unnatural backbones can sometimes form new favorable contacts not observed in the parent protein.
In order to modify the sheet of GB1 with unnatural backbone elements, we first attempted sequence-guided α→β residue replacement by 2:1 (αα→β) or 2:2 (αα→ββ) substitution strategies that we previously demonstrated in shorter peptide hairpins;12 however, these GB1 analogues showed no evidence of tertiary folding (Figure S3). As a more conservative modification, we incorporated N-methyl-α-residues13 at solvent-exposed positions on the two terminal strands of the four-stranded β-sheet. The resulting protein (4) yielded a nativelike CD scan and a Tm slightly below wild-type 1 upon thermal denaturation (Figure 4, Table 1). The destabilized folded state, somewhat surprising given the subtle nature of the modification, may result from altered solvation of the more hydrophobic backbone and/or isomerization about the new tertiary amide bonds.
GB1 has two turns connecting adjacent strands in the β-sheet. We employed two different backbone modifications, both known to promote turns in short peptides,14 to the larger protein: stereochemical inversion in the form of D-Pro (p) and Cα-methylation in the form of aminoisobutyric acid (Aib, U). We prepared GB1 analogue 5 (Figure 1) with Aib-Gly in place of the N-terminal β-turn of GB1 and d-Pro-Gly replacing four-residues of its C-terminal π-turn. d-Pro-Gly was used in the C-terminal hairpin based on its successful application in short peptides derived from this region of GB1.12a We hypothesized that the achiral Aib-Gly turn replacement in the N-terminal hairpin would allow the local protein environment to dictate the turn type and result in a native-like structure. The CD scan of 5 suggests a similar fold as wild type, but the modified protein showed a lower Tm (Figure 2, Table 1). A crystal structure of 5, refined to 2.0 Å resolution (Figure 3, Table S1), confirms the modified turns do not significantly perturb the tertiary fold; however, the structure also revealed possible factors contributing to the lower stability of 5 relative to 1. First, one of two chains in the asymmetric unit of 5 has locally elevated B-factors in the Aib-Gly turn and the loop that packs against it (Figure S7). Second, the Aib-Gly dipeptide formed a type-1′ turn type (Figure 4D), the mirror image of the corresponding type-1 turn in GB1.
Having demonstrated that the helix (2), loop (3), sheet (4), and turn (5) secondary structures of GB1 could be individually modified by introduction of various replacements for natural l-α-residues, we next tested whether these modifications could be performed simultaneously We combined the backbone alterations from 2, 3, 4, and 5 to generate GB1 analogue 6 (Figure 1). Approximately 20% of the backbone of 6 is composed of unnatural building blocks, but the sequence of side chains is almost identical to that of the parent protein GB1 (1). The CD signature observed for 6 was similar in shape to that of the helix-variant 2 but lower in magnitude (Figure 2A). A cooperative loss of the CD signal at 220 nm was seen as a function of temperature (Figure 2B, Table 1), although a fully folded baseline was not observed. The above two observations provide some evidence for a tertiary fold, but the folded state is clearly much less stable than the parent protein. This low stability is reasonable based on the thermodynamic data for 1-5. Assuming a simple additive loss of folding free energy from individual backbone modifications in 2-5 relative to wild-type protein 1, the predicted Tm for 6 would be ∼38 °C.
The above data suggest backbone modifications can be made in combination with roughly additive changes to tertiary folding free energy. A key question remaining was the degree to which the two sequences (side chain and backbone) in 6 were interrelated. Stated another way, would mutation of side chains on natural backbone 1 have the same effect when made on unnatural backbone 6? Prior work to improve the folded stability of GB1 through computational optimization of sequence generated a mutant (“NuG”, 7) with a tertiary fold identical to wild-type protein but significantly greater thermal stability (ΔΔGfold −5 kcal mol-1 for 7 vs. 1).15 We combined the side-chain sequence from 7 with the backbone sequence from 6 to generate protein 8 (Figure 1). The CD signature of 8 is similar in shape to that of 6, but 8 shows a cooperative unfolding transition at a much higher temperature (Figure 2, Table 1). The difference in folding free energy between 6 and 8 is substantial, and the Tm observed for 8 is close to wild-type GB1. The slight difference in folding cooperativity between 8 and 1 likely results from an altered folding pathway, noted previously for protein 7.15
A cooperative thermal unfolding transition is a common feature of tertiary folds;16 however, this observation alone is weak evidence of 6 or 8 showing a native-like structure. X-ray diffraction would provide a conclusive test of the nature of the folded state, but efforts to obtain diffraction-quality crystals of 6 or 8 have been unsuccessful to date. In the absence of such data, compelling evidence remains in the form of the similar stabilizing effect of identical side-chain alterations made to wild-type protein 1 and backbone-modified analogue 6. Collectively, the biophysical data obtained support the idea of the orthogonality of backbone and side-chain sequences.
In summary, we have demonstrated that a protein sequence encoding a compact tertiary fold can be used to design a heterogeneous backbone with native-like folding behavior. Helix, loops, sheet, and turns can be individually modified without compromising sequence-encoded folding, and alterations can be combined with near-additive effect on folding energetics. The unnatural building blocks examined here were all slightly destabilizing to the fold. The thermodynamics of residue substitutions will dictate how much of a protein can be altered while retaining the native folded state. The fraction of unnatural residues, in turn, will influence stability to enzymatic degradation, one motivation for the development of foldamer mimics of biomacromolecules. The designs reported here are meant to illustrate the general idea that proteins can be thought of as having mutually orthogonal backbone and side-chain sequences. Refined backbone modification strategies that make use of the full array of building blocks possible in peptidic oligomers may lead to species with fold and/or physiological stability improved over a prototype protein. Important open questions for future work include whether the design principles reported will be generalizable to different tertiary folds as well the relationship between backbone composition and proteolytic susceptibility.
Supplementary Material
Acknowledgments
Funding for this work was provided by the University of Pittsburgh and the National Institutes of Health (R01GM107161).
Footnotes
Supporting Information: Figures S1-S7, Tables S1-S2, and experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org. Coordinates and structure factors for 2, 3, and 5 have been deposited in the PDB under accession codes 4KGR, 4KGS, and 4KGT.
References
- 1.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.(a) Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]; (b) Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev. 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]; (c) Bautista AD, Craig CJ, Harker EA, Schepartz A. Curr Opin Chem Biol. 2007;11:685–692. doi: 10.1016/j.cbpa.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Horne WS. Expert Opin Drug Discov. 2011;6:1247–1262. doi: 10.1517/17460441.2011.632002. [DOI] [PubMed] [Google Scholar]; (f) Guichard G, Huc I. Chem Commun. 2011;47:5933–5941. doi: 10.1039/c1cc11137j. [DOI] [PubMed] [Google Scholar]
- 3.Wells JA, McClendon CL. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 4.(a) Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J Am Chem Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Horne WS, Price JL, Keck JL, Gellman SH. J Am Chem Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]
- 5.(a) Lee BC, Chu TK, Dill KA, Zuckermann RN. J Am Chem Soc. 2008;130:8847–8855. doi: 10.1021/ja802125x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Petersson EJ, Schepartz A. J Am Chem Soc. 2008;130:821–823. doi: 10.1021/ja077245x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Price JL, Hadley EB, Steinkruger JD, Gellman SH. Angew Chem Int Ed. 2010;49:368–371. doi: 10.1002/anie.200904714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase HS, Peterson-Kaufman KJ, Lan Levengood SK, Checco JW, Murphy WL, Gellman SH. J Am Chem Soc. 2012;134:7652–7655. doi: 10.1021/ja302469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne WS, Gellman SH. Acc Chem Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Lu W, Qasim MA, Laskowski M, Kent SBH. Biochemistry. 1997;36:673–679. doi: 10.1021/bi9625612. [DOI] [PubMed] [Google Scholar]; (b) Chapman E, Thorson JS, Schultz PG. J Am Chem Soc. 1997;119:7151–7152. [Google Scholar]; (c) Viles JH, Patel SU, Mitchell JBO, Moody CM, Justice DE, Uppenbrink J, Doyle PM, Harris CJ, Sadler PJ, Thornton JM. J Mol Biol. 1998;279:973–986. doi: 10.1006/jmbi.1998.1764. [DOI] [PubMed] [Google Scholar]; (d) Odaert B, Jean F, Melnyk O, Tartar A, Lippens G, Boutillon C, Buisine E. Protein Sci. 1999;8:2773–2783. doi: 10.1110/ps.8.12.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Arnold U, Hinderaker MP, Nilsson BL, Huck BR, Gellman SH, Raines RT. J Am Chem Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]; (f) Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]; (g) Horne WS, Yadav MK, Stout CD, Ghadiri MR. J Am Chem Soc. 2004;126:15366–15367. doi: 10.1021/ja0450408. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) David R, Gunther R, Baumann L, Luhmann T, Seebach D, Hofmann HJ, Beck-Sickinger AG. J Am Chem Soc. 2008;130:15311–15317. doi: 10.1021/ja802453x. [DOI] [PubMed] [Google Scholar]; (i) Lee BC, Zuckermann RN. ACS Chem Biol. 2011;6:1367–1374. doi: 10.1021/cb200300w. [DOI] [PubMed] [Google Scholar]; (j) Valverde IE, Lecaille F, Lalmanach G, Aucagne V, Delmas AF. Angew Chem Int Ed. 2012;51:718–722. doi: 10.1002/anie.201107222. [DOI] [PubMed] [Google Scholar]
- 9.(a) Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]; (b) Frericks Schmidt HL, Sperling LJ, Gao YG, Wylie BJ, Boettcher JM, Wilson SR, Rienstra CM. J Phys Chem B. 2007;111:14362–14369. doi: 10.1021/jp075531p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horne WS, Price JL, Gellman SH. Proc Natl Acad Sci USA. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc Natl Acad Sci USA. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinert ZE, Musselman ED, Elcock AH, Horne WS. ChemBioChem. 2012;13:1107–1111. doi: 10.1002/cbic.201200200. [DOI] [PubMed] [Google Scholar]
- 12.(a) Lengyel GA, Frank RC, Horne WS. J Am Chem Soc. 2011;133:4246–4249. doi: 10.1021/ja2002346. [DOI] [PubMed] [Google Scholar]; (b) Lengyel GA, Horne WS. J Am Chem Soc. 2012;134:15906–15913. doi: 10.1021/ja306311r. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee J, Rechenmacher F, Kessler H. Angew Chem Int Ed. 2013;52:254–269. doi: 10.1002/anie.201205674. [DOI] [PubMed] [Google Scholar]
- 14.(a) Haque TS, Little JC, Gellman SH. J Am Chem Soc. 1996;118:6975–6985. [Google Scholar]; (b) Masterson LR, Etienne MA, Porcelli F, Barany G, Hammer RP, Veglia G. Biopolymers. 2007;88:746–753. doi: 10.1002/bip.20738. [DOI] [PubMed] [Google Scholar]
- 15.Nauli S, Kuhlman B, Le Trong I, Stenkamp RE, Teller D, Baker D. Protein Sci. 2002;11:2924–2931. doi: 10.1110/ps.0216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betz SF, Raleigh DP, Degrado WF. Curr Opin Struct Biol. 1993;3:601–610. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.