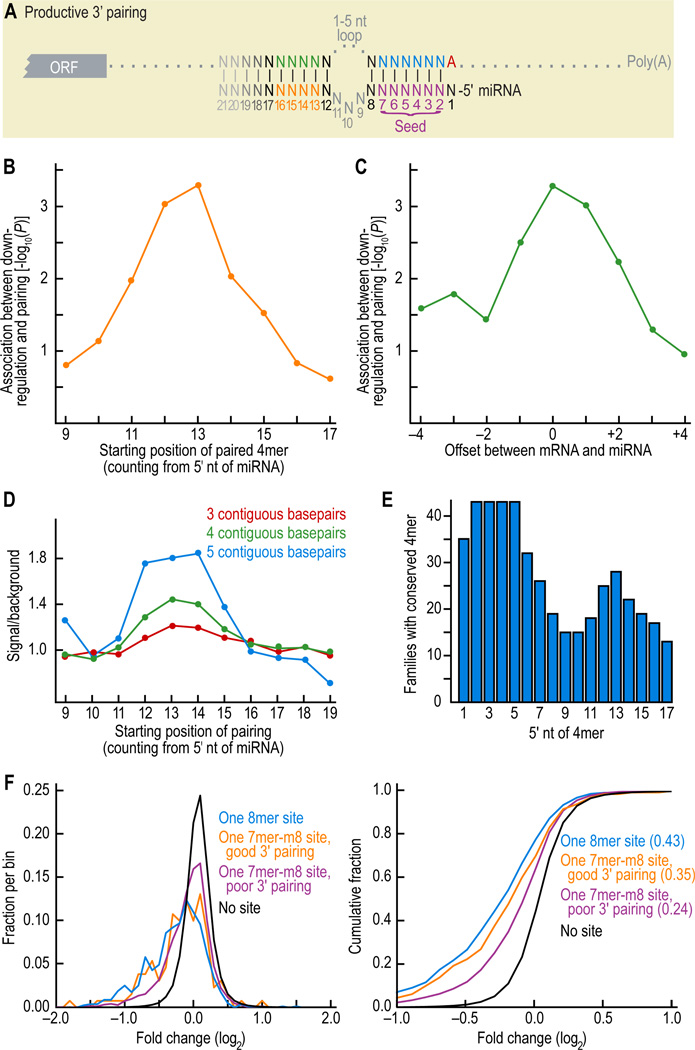
Figure 2. Characteristics of beneficial 3' pairing.
(A) Pairing scheme, highlighting the core region of productive 3' pairing between the miRNA (orange) and 3'UTR (green).
(B) Preferential position of supplementary pairing within the miRNA 3' region. Starting with 7mer-m8 sites for representative vertebrate miRNAs of Table S1, a 4mer window was slid across the 3' end of the miRNA, searching for its Watson-Crick match in the opposing region of the message. A 3' pairing score was assigned to each miRNA 4mer, crediting one point for each contiguous pair within the 4mer, a half point for extending the contiguous pairing 1 nt upstream and a half point for extending it 1 nt downstream. The position of the miRNA 4mer and its complement in the message were allowed to be offset, but a ½ point penalty was assessed for each nucleotide of offset beyond ± 2 nt, and pairing to message positions already paired to the miRNA seed region (1–8) was disallowed. When the 4mer had alternative regions of contiguous pairing, it was assigned the highest of the alternative scores. For each position, the Spearman correlation between the score and down-regulation on the array was determined and its P value plotted.
(C) Preferred offsets between paired regions of miRNA and mRNA. Analysis of (B) was repeated using a 4mer fixed at miRNA nucleotides 13–16, and the correlation between score and down-regulation was evaluated for alternative pairing offsets. A positive offset shifts the miRNA 3' pairing segment to the right, relative to the message.
(D) Preferential positions of conserved pairing. Human 3'UTRs were scanned for sites with at least a 6mer seed match to miRNAs of Table S1. For each contiguous 4mer beginning at nucleotide 9 of the miRNA, we searched for the complementary Watson-Crick 4mer directly opposite in the message, allowing for a ± 2-nt offset and excluding overlap into nucleotides 1–8. Those cases in which the seed and its supplemental 4mer were co-conserved were considered conserved instances of 3' pairing. This was compared to control chimeric miRNA sequences with the same 5' seed sequence but with the 3' end of a different miRNA, and signal/background was calculated. The analysis was repeated for 3mers and 5mers.
(E) 4mers conserved among paralogous human miRNAs. For each position the number of human miRNA families that have a perfectly conserved 4mer is indicated; families with only a single human paralog were excluded.
(F) Effectiveness of 7mer-m8 sites compensated with either good or poor 3' pairing. Distributions (left) and cumulative distributions (right) of changes in abundance of mRNAs following miRNA transfection were monitored with microarrays, and are displayed as in Figure 1B, including for reference the results of canonical 8mer sites. Sites with good pairing were those with scores ≥ 4 and sites with poor pairing were those with scores ≤ 2. Pairing score was determined as in (B) but using a fixed miRNA 4mer corresponding to nucleotides 13–16 and crediting contiguous pairing elsewhere with ½ point per pair. Sites with good pairing were significantly more effective than sites with poor pairing (P = 0.007, 1-sided K-S test).