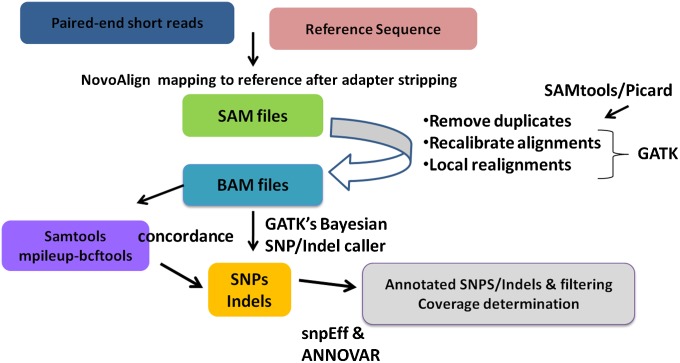
Fig. 1.
Workflow for processing NGS data. Raw sequencing data are aligned against the reference sequence using Novoalign software from NovoCraft. SAM files are preprocessed using SAMtools and Picard to create BAM files and remove duplicates. The Genome Analysis Toolkit (GATK) is then used to recalibrate the alignments, perform local realignments, and identify SNPs and indels. Finally, SnpEff and ANNOVAR are used to annotate variants.