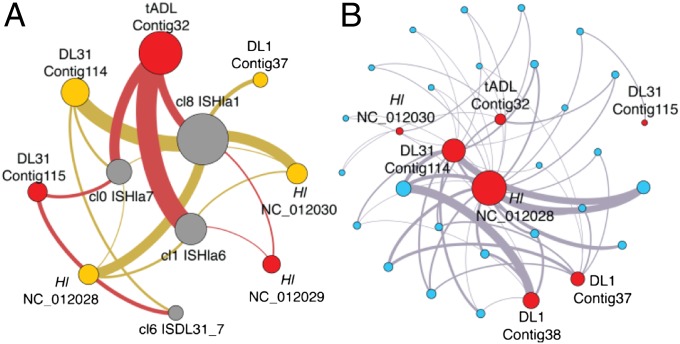
Fig. 3.
Association networks for DL features. Bipartite networks for features identified within the four DL haloarchaeal genomes, using Fruchterman–Reingold layout. (A) Nonredundant sequences identified from short highly degenerate regions (gray nodes; read depth ∼2,000) and their containing replicons (red nodes primary replicons, orange nodes secondary replicons), where node radius scales with weighted degree and edge weights are proportional to degenerate region frequency. For clarity, the figure has been filtered to exclude weak (low weight) edges (e.g., between DL1 and cl0 and cl6). (B) HIR (identity > 99.8%, length > 5 kb) (blue nodes) and their containing replicon (red nodes) where node radius scales linearly with weighted degree, and edge weights are proportional to region length.