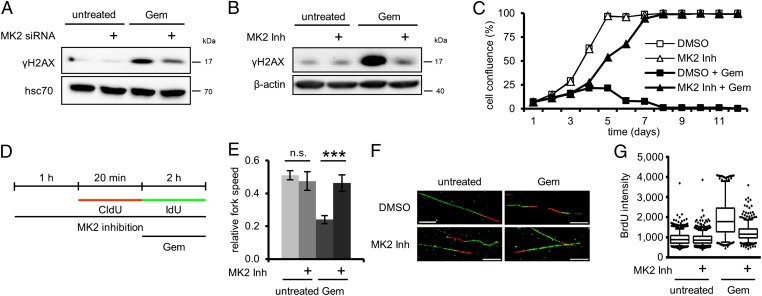
Fig. 2.
Gemcitabine-induced H2AX phosphorylation and reduced replication fork speed depend on MK2. (A and B) H2AX phosphorylation following gemcitabine treatment requires MK2. (A) Cells were depleted of MK2 and treated with 100 nM gemcitabine for 20 h or left untreated. Cell lysates were analyzed by immunoblotting. (B) Cells were treated with 100 nM gemcitabine for 24 h or left untreated and simultaneously treated with MK2 inhibitor or DMSO, followed by immunoblot analysis. (C) Cell survival after gemcitabine treatment is improved by MK2 inhibition. On day 1, cells were treated with 100 nM gemcitabine and MK2 inhibitor or DMSO for 24 h. Cell confluence was determined daily for 12 d by light microscopy. (D–F) MK2 inhibition rescues reduced replication fork speed caused by gemcitabine. (D) Labeling protocol for DNA fiber analysis of replication fork speed. Cells were pretreated with MK2 inhibitor or DMSO for 1 h and throughout the experiment. Cells were then pulse labeled with 5-chloro-2′-deoxyuridine (CldU) for 20 min, followed by IdU for 2 h and simultaneous exposure to 400 nM gemcitabine. CldU and IdU were detected by using specific primary antibodies and secondary antibodies in red and green, respectively. (E) Average relative replication fork speed (ratio of length of IdU-labeled vs. length of CldU-labeled tracks) in cells treated as in D in dependence of gemcitabine and MK2 inhibition (n = 3; ***P = 0.0002). (F) Representative images of fibers treated as in D. (G) MK2 inhibition reduces ssDNA accumulation upon gemcitabine treatment. BrdU-labeled cells were treated with 300 nM gemcitabine and MK2 inhibitor or DMSO for 24 h. Cells were fixed and processed for ssDNA quantification by immunofluorescent detection of accessible BrdU without DNA-denaturing treatment. Image is representative of three independent replicates.