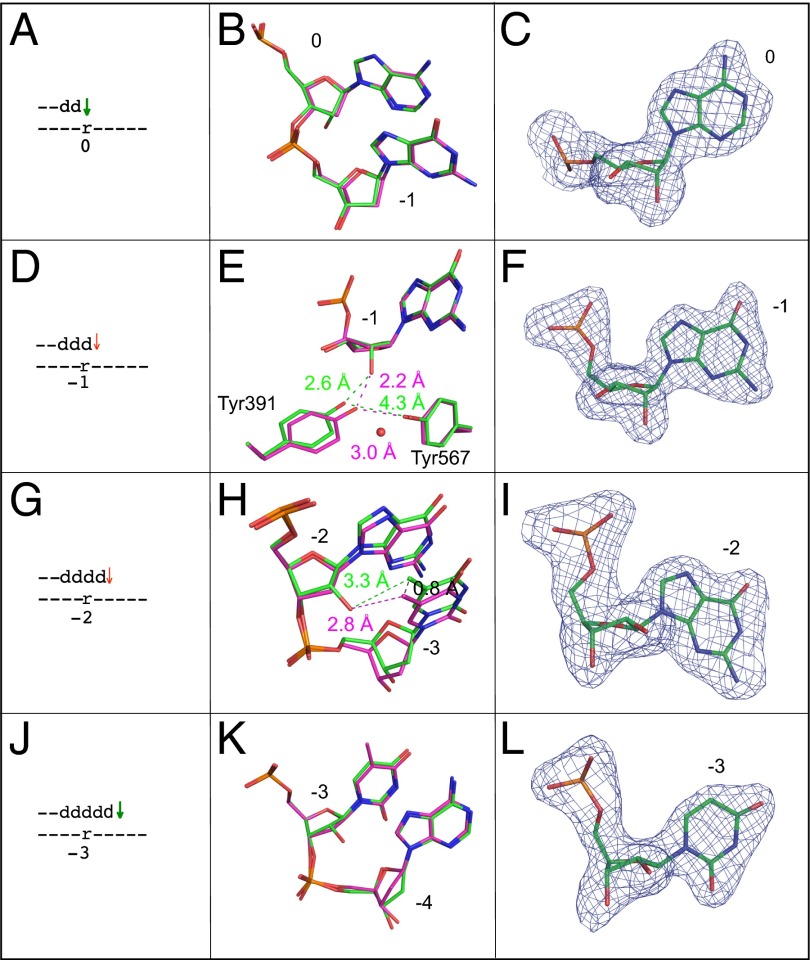
Fig. 2.
Superposition of all-DNA structure (magenta) with four different single ribonucleotide structures (green). (A) Schematic depicting the ribonucleotide in the nascent base pair binding pocket (position 0). A green arrow implies that dNTP insertion is not strongly reduced, as inferred from the relatively normal termination after the preceding incorporation (Fig. 1 D1 and D2, −1-dG). (B) Overlay of the ribonucleotide-containing structure depicted in A with the all-DNA structure, showing template positions 0 and −1. (C) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide in the nascent base pair binding pocket. (D) Stick diagram depicting the location of ribonucleotide in the primer-terminal base pair (−1 position). The red arrow implies that dNTP insertion at this position (or possibly translocation before insertion) is problematic, as indicated by the increased termination following insertion opposite rA (Fig. 1 D1 and D2, rA). (E) Overlay of the ribonucleotide containing structure depicted in D with the all-DNA structure, showing template position −1, Tyr391, and Tyr567. The red sphere indicates the position of an additional water molecule in the ribonucleotide-containing structure. (F) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide in the −1 position. (G) Stick diagram of the position of the ribonucleotide located 2 bp upstream of the active site (−2 position). As above, the red arrow implies that dNTP insertion at this position is problematic (Fig. 1 D1 and D2, +1-dC). (H) Overlay of the ribonucleotide containing structure with the all-DNA structure at template positions −2 and −3. (I) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide at position −2. (J) Stick diagram of the position of ribonucleotide located 3 bp upstream of the active site (−3 position). As explained above, the green arrow indicates that insertion is normal (Fig. 1 D1 and D2, +2dA). (K) Overlay of the ribonucleotide-containing structure with the all-DNA structure at template positions −3 and −4. (L) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide at position −3.