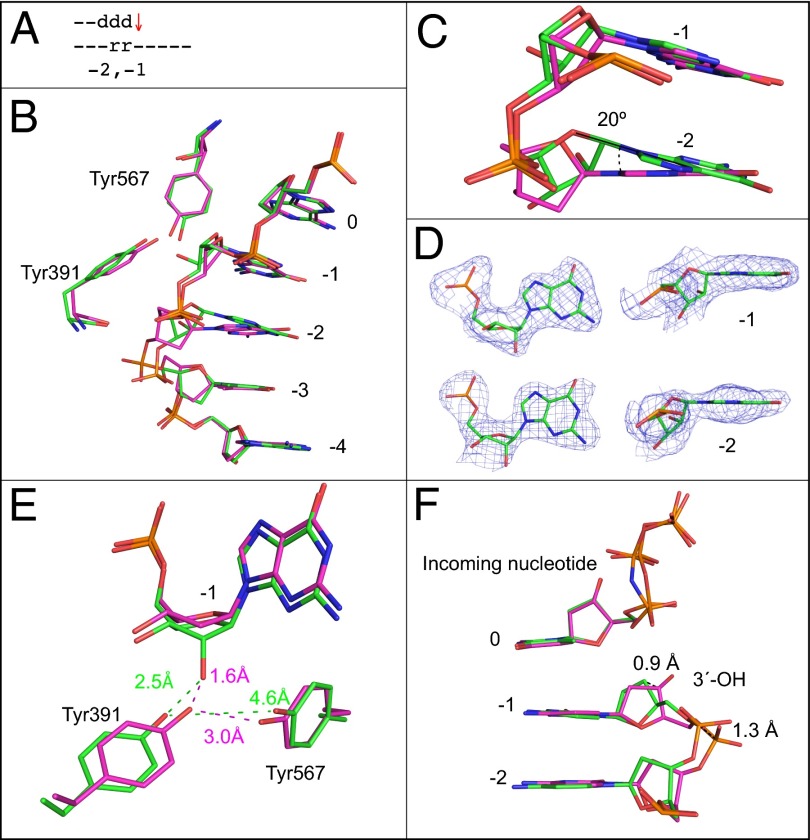
Fig. 4.
Superposition of the all-DNA structure (magenta) with the structure containing ribonucleotides at −1 and −2 positions (green). (A) Schematic indicating the positions of ribonucleotides at −1 and −2. The red arrow indicates that termination frequency is increased ∼10-fold compared with bypass of an all-DNA template, (Fig. 1D3, 5′-rA). (B) Superposition of template bases at 0 to −4 positions, Tyr391, and Tyr567. (C) Overlay of ribonucleotides at the −1 and −2 position with the all-DNA structure. (D) Simulated annealing Fo-Fc omit maps contoured at 3 σ are shown in blue for the two ribonucleotides at the −1 and −2 positions. (E) Overlay with the all-DNA structure showing Tyr391, Tyr567, and the nucleotide at the −1 position. (F) Overlay of primer terminus and incoming nucleotide with the all-DNA structure.