Significance
An important part of the response to invading pathogens is activation of the transcription factor STAT3 in response to IL-6. This normal response is terminated rapidly by the major negative regulator suppressor of cytokine signaling 3 (SOCS3). Abnormal prolonged activation of STAT3 contributes to the pathology of cancer and to chronic inflammatory diseases. We show that prolonged activation of STAT3 results from association of the IL-6 receptor with the epidermal growth factor receptor, which in turn activates STAT3 in a way that is not inhibited by SOCS3. Prolonged STAT3 activation supports sustained expression of a subset of proteins that play important roles in inflammation and in cancer, where both IL-6 secretion and epidermal growth factor receptor levels are often elevated.
Abstract
The activation of STAT3 by tyrosine phosphorylation, essential for normal development and for a normal inflammatory response to invading pathogens, is kept in check by negative regulators. Abnormal constitutive activation of STAT3, which contributes to the pathology of cancer and to chronic inflammatory diseases such as rheumatoid arthritis, occurs when negative regulation is not fully effective. SOCS3, the major negative regulator of STAT3, is induced by tyrosine-phosphorylated STAT3 and terminates STAT3 phosphorylation about 2 h after initial exposure of cells to members of the IL-6 family of cytokines by binding cooperatively to the common receptor subunit gp130 and JAKs 1 and 2. We show here that when the epidermal growth factor receptor (EGFR) is present and active, STAT3 is rephosphorylated about 4 h after exposure of cells to IL-6 or oncostatin M and remains active for many hours. Newly synthesized IL-6 drives association of the IL-6 receptor and gp130 with EGFR, leading to EGFR-dependent rephosphorylation of STAT3, which is not inhibited by the continued presence of SOCS3. This second wave of STAT3 activation supports sustained expression of a subset of IL-6-induced proteins, several of which play important roles in inflammation and cancer, in which both IL-6 secretion and EGFR levels are often elevated.
After ligand-induced dimerization of the IL-6 receptor (IL-6R), the associated kinases JAK1 and JAK2 cross-phosphorylate tyrosine residues of adjacent glycoprotein 130 (gp130) subunits of the complex. The SH2 domain of STAT3 binds to newly phosphorylated tyrosines, followed by the phosphorylation of Y705 of STAT3 (1). Two phosphorylated STAT3 monomers then dimerize and translocate to the nucleus, where they activate the transcription of many downstream genes whose products mediate the diverse effects of STAT3 in development and disease (2). Suppressor of cytokine signaling 3 (SOCS3), the major negative regulator of IL-6-dependent signaling, is highly induced by activated STAT3, strongly inhibiting further STAT3 phosphorylation and thus collaborating with additional negative regulatory mechanisms to prevent excessive activation of potentially deleterious gene expression (3). SOCS3 down-regulates the phosphorylation of STAT3 about 90 min after an initial exposure to IL-6 (4), using two opposing surfaces to bind to JAK2 and gp130 simultaneously (5). However, even in the continued presence of SOCS3, STAT3 is rephosphorylated in response to IL-6 after about 4 h through a mechanism that has been obscure until now. Rephosphorylation of STAT3 might occur because the interaction between SOCS3 and the IL-6 receptor is somehow prevented or because continued STAT3 phosphorylation occurs through a mechanism that is immune to SOCS3. The epidermal growth factor receptor (EGFR) is well known to catalyze the tyrosine phosphorylation of STAT3 in response to EGF (6), and the intrinsic kinase activity of the receptor, but not any of the JAKs, is required for this reaction (7). On activation by EGF, the EGFR dimerizes to facilitate cross-phosphorylation of several tyrosine residues, including Y1068, the binding site for STAT3 (8). Multiple tyrosine phosphorylations of EGFR initiate downstream signaling cascades that facilitate proliferation, regeneration, and tumorigenesis (9).
During the early phase of mouse development, IL-6 mRNA is differentially expressed and translated into the mature protein (10), indicating the involvement of IL-6 in the growth and differentiation of the preimplantation mouse embryo (11). One of the most notable activities of the IL-6 family of cytokines, from the viewpoint of embryonic development, is the expression of the IL-6 family member leukemia inhibitory factor (LIF) in the murine uterine endometrial glands of pregnant mice, coincident with the onset of blastocyst implantation (12, 13). Mice in which the IL-6 or LIF genes have been knocked out show much less severe defects than might have been anticipated from the widespread effects of these cytokines (14, 15), perhaps reflecting the shared use of common gp130 signaling pathways by other gp130-dependent cytokines. In addition, intriguingly, mice lacking SOCS3 die between days 11 and 13 of gestation (16), showing the importance of down-regulation of STAT3 activation in development.
The STAT transcription factors, and especially STAT3, play vital roles in tumor initiation and progression (17, 18). Aberrant activation of STAT3 leads to the overexpression of oncogenes that drive proliferation and metastasis and inhibit apoptosis (19). As an activator of the JAK-STAT signaling pathway, IL-6 can act in both a proinflammatory and anti-inflammatory manner. Deregulation of IL-6 production can cause severe disease, such as rheumatoid arthritis, psoriasis, arteriosclerosis, and cancer (20, 21). Through the JAK/STAT signaling pathway, IL-6 mediates the production of IL-17 and IL-10 while regulating TH17, which is important in autoimmunity (22). IL-6 elevation and the chronic inflammation that follows are protumorigenic. IL-6 exerts powerful antiapoptotic functions through the induction of B-cell lymphoma 2 (BCL-2), B-cell lymphoma-extra large (BCL-xL), and related inhibitors (23). Therefore, anti–IL-6/IL-6R monoclonal antibodies are used to inhibit aberrant and prolonged IL-6 signaling in the treatment of autoimmunity and cancer (21). Because EGFR activation can mediate both IL-6 production (24, 25) and STAT3 activation, we investigated the role of EGFR in regulating STAT3 activation in response to IL-6.
We confirm the observation of others (3, 26) that, after the initial phosphorylation of STAT3 in response to IL-6 and subsequent inhibition by SOCS3, a second wave of activation leads to the rephosphorylation of STAT3. We now find that rephosphorylation requires new IL-6 synthesis and secretion, leading to ligand-dependent association of IL-6R and EGFR. STAT3 phosphorylation continues to be driven for many hours by this two-receptor complex, which is immune to inhibition by SOCS3, thus increasing and prolonging the expression of many STAT3-dependent proteins that are involved in tumorigenesis and immune regulation.
Results
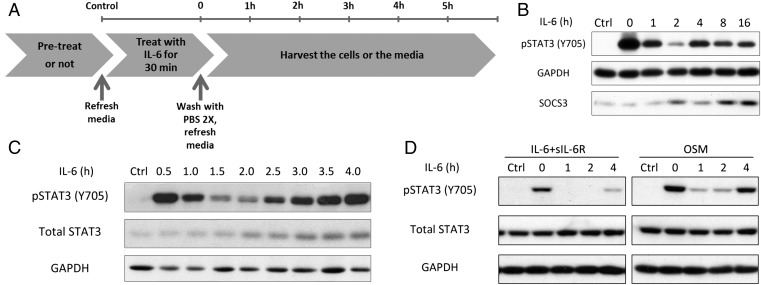
IL-6 Signaling Is Prolonged in a Biphasic Pattern.
To examine the second wave of STAT3 activation in detail, we treated DLD1 (colon cancer cells) with IL-6 for 30 min, followed by washout, and measured STAT3 phosphorylation as a function of time (Fig. 1 A and B). Phosphorylation lasts for many hours, with a second wave of STAT3 activation apparent 4 h after IL-6 has been removed. This second wave of STAT3 activation was also observed in a similar experiment in which additional early times were analyzed (Fig. 1C). 2fTGH (adenocarcinoma cells) exposed to IL-6 show a weaker but still significant second wave at 4 h (Fig. 1D). These cells respond more strongly to oncostatin M (OSM), another cytokine that uses the gp130 common receptor subunit, with a clear second wave of STAT3 activation (Fig. 1D).
Fig. 1.
IL-6 signaling is prolonged in a biphasic pattern. (A) Flowchart of the washout procedure. (B) DLD1 colon cancer cells were treated with IL-6 (100 ng/mL) for 30 min, the cells were washed with PBS, and the medium was replaced. (C) The experiment is similar to B, but with more early times. (D) 2fTGH cells were treated with IL-6 (100 ng/mL) and with soluble IL-6R (125 ng/mL, included because 2fTGH cells do not have a high level of endogenous IL-6R) or OSM (10 ng/mL) for 30 min, followed by washing and replacement of the medium. Each experiment in B–D was carried out independently between two and six times, with results similar to the representative examples that are shown.
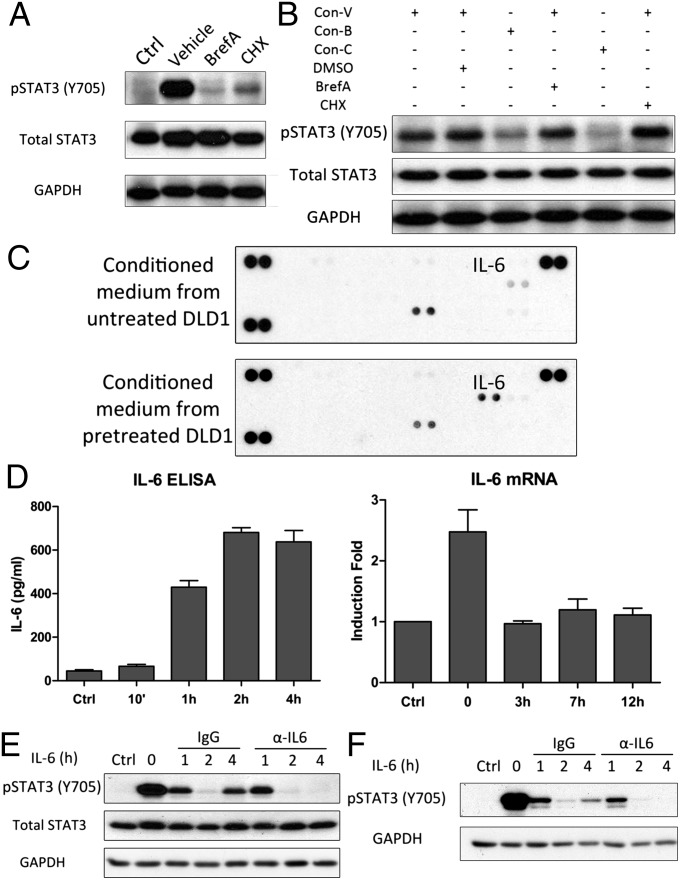
The Biphasic Pattern Is Mediated by New IL-6 Synthesis and Secretion.
To determine whether IL-6–treated cells secrete any factor that induces the biphasic pattern, we pretreated the cells with cycloheximide or brefeldin A to block newly induced protein synthesis and secretion, respectively. Both agents caused strong inhibition of the biphasic pattern (Fig. 2A). Conditioned medium from IL-6–treated cells induces STAT3 phosphorylation, but not if the cells are pretreated with cycloheximide or brefeldin A (Fig. 2B). However, the ability of conditioned medium from control IL-6–treated cells to activate STAT3 was not inhibited by cycloheximide or brefeldin A added later (Fig. 2B). We conclude that factors that are newly synthesized and secreted mediate the second wave. We used a cytokine array to identify such factors by analyzing media conditioned for 4 h after washout from cells treated with IL-6 for 30 min, finding that IL-6 is the dominant induced factor (Fig. 2C). The supernatant media were also analyzed by ELISA to quantify IL-6 production. The lack of IL-6 in media collected 10 min after washout shows that the removal of the IL-6 added initially is efficient (Fig. 2D, Left). IL-6 was secreted by 1 h after washout and remained in the media at a similar level for at least 4 h (Fig. 2D, Left). An analysis by real-time PCR showed that IL-6 mRNA was induced within the first 30 min of initial treatment with IL-6 and then declined (Fig. 2D, Right). An antibody against human IL-6 totally ablated the second wave of STAT3 activation in DLD1 cells, showing that no other secreted protein is responsible (Fig. 2E). The same phenomenon was observed in HepG2 cells (Fig. 2F). In summary, we show that treatment with IL-6 induces the synthesis and secretion of more IL-6 and that this induction is necessary for the second wave of STAT3 activation.
Fig. 2.
The biphasic pattern is mediated by IL-6 secretion. (A) DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min, followed by washing and replacement of the medium with or without cycloheximide (CHX; 1 μg/mL) or brefeldin A (BrefA; 1 μg/mL). The cells were harvested 4 h later. (B) The conditioned media from A were collected 4 h after washout and used to treat fresh DLD1 cells, with or without inhibitors. The cells were harvested after 1 h. BrefA, brefeldin A; Con-B, conditioned medium from cells treated with BrefA (1 μg/mL); Con-C, conditioned medium from cells treated with CHX (1 μg/mL); Con-V, conditioned medium from cells without inhibitor; CHX, cycloheximide; V, vehicle. (C) Conditioned medium from A, collected 4 h after washout, was analyzed, using the R&D Systems Human Cytokine Array, Panel A. (D) DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min, followed by washing and replacement of the medium. The medium was analyzed by ELISA at the times indicated (Left). IL-6 mRNA was analyzed by real-time PCR (Right). Values are the means ± SD from three independent experiments. (E) DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min, followed by washing and replacement of the media with control rabbit IgG or anti-human IL-6. (F) HepG2 cells were treated with IL-6 (100 ng/mL) for 30 min, followed by washing and replacement of the media with control rabbit IgG or anti-human IL-6. Each experiment in A, B, E, and F was carried out independently between two and six times, with results similar to the representative examples that are shown.
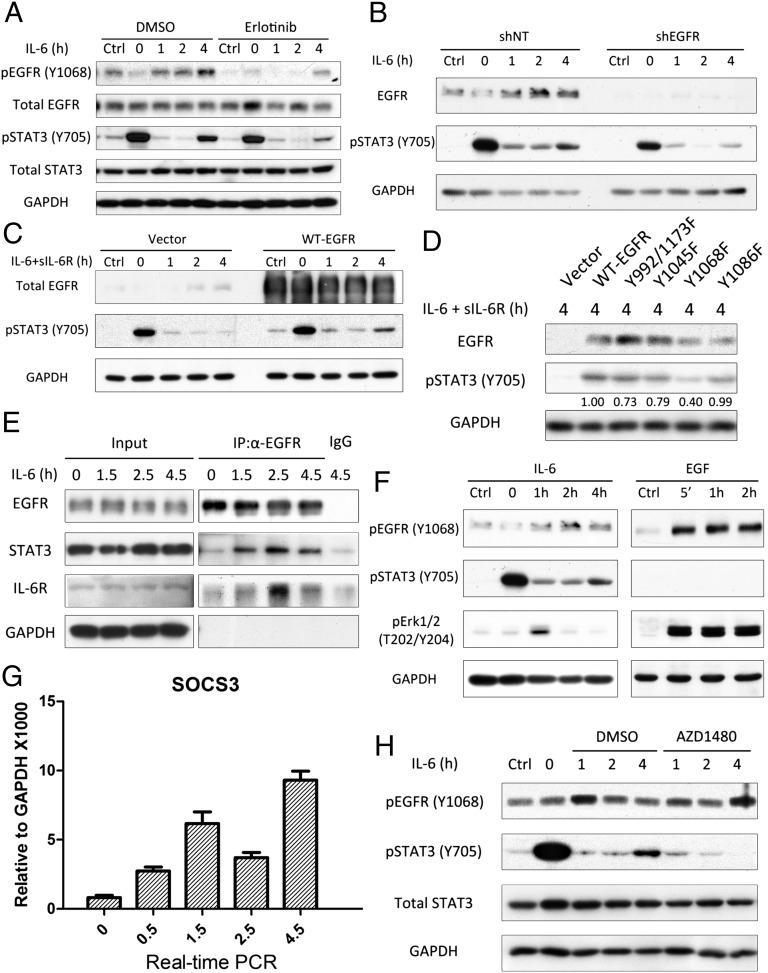
EGFR Is Required for the Second Wave of STAT3 Phosphorylation.
EGFR is also known to activate STAT3 by phosphorylating Y705 (24, 25). To study whether EGFR is involved in the second wave of STAT3 activation, we used the well-known EGFR inhibitor erlotinib, which, as expected, blocks EGFR Y1068 phosphorylation (Fig. 3A) and also inhibits the second wave of STAT3 phosphorylation (Fig. 3A). To obtain further evidence for the involvement of EGFR, we knocked its expression down in DLD1 cells, observing a substantial reduction of the second wave (Fig. 3B). We also observed very little STAT3 phosphorylation in the second wave in HEK 293T cells, which express a low level of EGFR (Fig. 3C, Left). Therefore, 293T cells provide an essentially null background in which the role of wild-type EGFR and the functions of various EGFR mutants can be evaluated. Overexpression of EGFR in 293T cells did enhance the second wave of STAT3 activation without affecting the initial response to IL-6 (Fig. 3C, Right). We also overexpressed several EGFR tyrosine to phenylalanine (Y-to-F) mutants (Y992/1173F, Y1045F, Y1068F, Y1086F) in the same cells to identify a tyrosine residue, the phosphorylation of which might be important in the second wave. Compared with wild-type EGFR and the other mutants, the second wave of STAT3 activation was reduced primarily in cells transfected with the Y1068F mutant (Fig. 3D). Because, after it has been phosphorylated in response to EGF, Y1068 of EGFR provides a docking site for the SH2 domain of STAT3 (27), this result suggests that the phosphorylation of Y1068 in the second wave (Fig. 3A) provides a potential docking site for STAT3, allowing it to be phosphorylated even in the presence of SOCS3. The interaction between EGFR and STAT3 in response to IL-6 was revealed by immunoprecipitation (Fig. 3E). The binding of EGFR to both STAT3 and IL-6R was increased in response to IL-6 treatment at the beginning of the second wave. However, the phosphorylation of ERK, a well-known target of EGFR, was activated by long-term treatment of DLD1 cells with IL-6 much more weakly than it was activated by the treatment of the same cells with EGF, showing clearly that IL-6-induced EGFR activation is distinct from the activation induced by EGF (Fig. 3F). SOCS3 is induced in two phases (Figs. 3G and 1B). The initial increase in SOCS3 is likely to be the major factor leading to down-regulation of STAT3 phosphorylation after 1.5 h. However, after 4.5 h, even higher levels of SOCS3 (Fig. 3G) no longer inhibit STAT3 phosphorylation. The JAK inhibitor AZD1480, added 30 min after IL-6 stimulation, did not affect the initial response but did block the second wave (Fig. 3H). This result is consistent with a model in which the association with EGFR of the IL-6 receptor, together with its tightly associated kinases JAK 1 and 2, allows one of these kinases to phosphorylate Y1068 of EGFR. Additional work will be needed to clarify the roles of JAK 1 and 2 in detail.
Fig. 3.
EGFR is crucial for the biphasic pattern. (A) DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min and washed twice with PBS before replacing the media. The cells were exposed to erlotinib (10 μM) for 30 min before treatment, and the erlotinib was left in during all subsequent steps. (B) DLD1 cells were transfected with scrambled shRNA (shNT; nontargeted) or shRNAs against EGFR and then selected with puromycin for 1 wk. The cells were then treated with IL-6 (100 ng/mL) for 30 min and washed twice before replacement of the media. (C and D) 293Tcells were transfected with pCDNA3.1 (Vector) or pCDNA3.1 expressing WT-EGFR or the Y992/1173F, Y1045F, Y1068F, or Y1086F mutant proteins. After 48 h, the cells were treated with IL-6 (100 ng/mL) and soluble IL-6R (125 ng/mL, included because 293T cells do not have a high level of endogenous IL-6R) for 30 min and washed twice with PBS, followed by replacement with normal media. In D, the numbers shown are the ratios of the intensities of the bands for pSTAT3, divided by the intensities for the bands for EGFR, normalized to 1.00 for WT-EGFR. (E) DLD1 cells were treated with 100 ng/mL of IL-6. Membrane-bound proteins were used for immunoprecipitation of EGFR. (F) DLD1 cells were treated with IL-6 (100 ng/mL) or EGF (5 ng/mL) without washout and then harvested after 4 h. Note that the loading (GAPDH) and exposure times (equivalent signals in the control lanes) are similar for the IL-6–treated and EGF-treated cells. (G) 293T cells were treated with IL-6 (100 ng/mL) and soluble IL-6R (125 ng/mL). Cells were harvested at the times indicated and analyzed by real-time PCR. Values are the means ± SD from three independent experiments. (H) DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min and washed twice with PBS before replacing the media, with or without AZD1480 (1 μM). Each of these experiments (except G) was carried out two independent times, with results similar to the representative examples that are shown.
Analysis of Gene Expression in Response to the Second Wave of STAT3 Phosphorylation.
We examined gene expression to help understand the biological significance of the second wave. A microarray analysis was carried out, using DLD1 cells treated with IL-6 for 30 min, followed by washout and replacement of the medium. The cells were collected for analysis 7 and 12 h later. STAT3 activation followed the expected biphasic pattern, and treatment with anti–IL-6 or knockdown of EGFR weakened the enhancement of STAT3 phosphorylation dramatically at late times (Fig. S1A). Of the 44,053 genes represented on the array, 1,960 were induced after the initial treatment with IL-6. The expression of 253 of these genes was inhibited by anti–IL-6 at late times (Table 1 and Table S1), showing that their expression is enhanced in the second wave. Analysis of the induction of several of these genes by real-time PCR confirmed the array results (Fig. S1 B and C). Interestingly, proteins encoded by many of the mRNAs whose expression is enhanced in the second wave are involved in major physiological effects of prolonged exposure to IL-6; for example, FBJ murine osteosarcoma viral oncogene homolog (FOS), Mucin 1, cell surface associated (MUC1), and ATP-binding cassette sub-family B member 1 (ABCB1) in cancer and interferon regulatory factor 1 (IRF1), interferon regulatory factor 8 (IRF8), and serum amyloid A1 (SAA1) in immune responses. Many of the genes affected by anti–IL-6 are also affected by loss of EGFR expression, consistent with the results of inhibiting the second wave with anti–IL-6. However, some gene expression inhibited by anti–IL-6 is not inhibited by EGFR knockdown (see CXCL2 and GADD45G in Fig. S1C and additional examples in Table 1), perhaps reflecting the incomplete knockdown shown in Fig. S1A.
Table 1.
Genes affected by the second wave of STAT3 activation
| Gene | Fold induction compared with untreated cells |
|||||
| 7 h | 7 h + Ab | 7 h − EGFR | 12 h | 12 h + Ab | 12 h − EGFR | |
| ABCB1 | 1.4 | 0.9 | 1.0 | 2.0 | 1.1 | 1.1 |
| CDK5RAP3 | 1.7 | 0.7 | 0.2 | 2.7 | 1.3 | 0.5 |
| FOS | 7.7 | 0.9 | 2.3 | 3.0 | 1.0 | 1.9 |
| GADD45G | 2.0 | 1.0 | 3.3 | 1.7 | 1.1 | 2.7 |
| HDAC4 | 1.4 | 1.2 | 1.6 | 2.4 | 1.4 | 1.9 |
| IGSF3 | 1.1 | 1.8 | 0.8 | 2.0 | 1.1 | 1.0 |
| IRAK1BP1 | 1.1 | 1.2 | 2.4 | 1.6 | 1.0 | 1.7 |
| IRF1 | 2.2 | 1.3 | 1.8 | 1.9 | 1.4 | 1.3 |
| IRF8 | 1.6 | 0.7 | 1.1 | 1.9 | 1.2 | 0.9 |
| MAPK8 | 1.8 | 0.8 | 6.0 | 1.2 | 1.0 | 6.8 |
| MLH3 | 2.3 | 0.4 | 0.1 | 1.6 | 1.1 | 0.6 |
| MUC1 | 3.5 | 3.5 | 2.3 | 4.9 | 1.9 | 1.9 |
| MUC6 | 1.3 | 0.8 | 1.3 | 1.8 | 0.6 | 0.6 |
| PRKAA1 | 1.7 | 1.0 | 0.9 | 1.6 | 1.5 | 1.0 |
| SAA1 | 2.4 | 1.2 | 11.4 | 2.6 | 1.6 | 15.3 |
See the legend to Fig. S1 for the protocol. DLD1 cells were treated with IL-6 for 30 min, followed by washout and replacement of the medium, with or without anti–IL-6. Cells in which EGFR was knocked down were also used, without exposure to anti-Il-6 antibody (Ab). Cells were collected for analysis 7 or 12 h after washout. The expression of 47,323 probes on an Illumina HumanHT-12 v4 Expression BeadChip was analyzed. The average signal for each probe was used to determine expression levels. Genes with average signals below 25 and detection P values greater than 0.01 in the treated cells were excluded from the analysis. The numbers in the table are fold inductions relative to untreated control cells. Inductions of less than 1.5-fold were not scored, and reductions of induction of less than twofold in response to anti–IL-6 were also not scored. SAA1 was not detected in untreated shEGFR cells but was induced in response to IL-6; the numbers are the fold inductions compared with the levels of expression in untreated shNT (nontargeting shRNA) cells.
Discussion
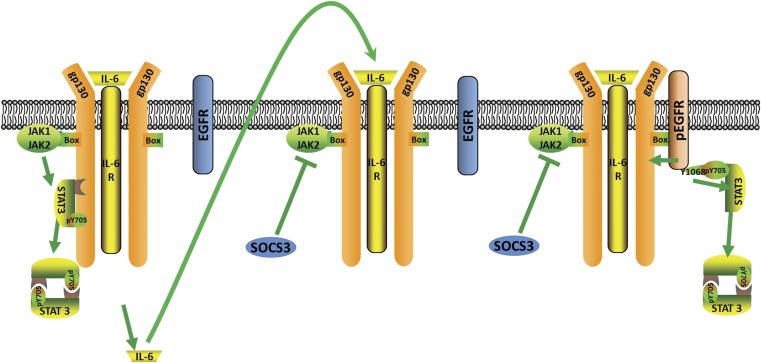
IL-6 has diverse roles in normal development, the acute phase response, chronic inflammation, autoimmunity, endothelial cell dysfunction, and cancer progression (21, 28), and IL-6 is a therapeutic target in several different human diseases, including diabetes, arthritis, and cancer. We now describe how cells can achieve a prolonged response to IL-6, overcoming the SOCS3-dependent inhibition of IL-6R (Fig. 4). The second wave of STAT3 activation involves IL-6-driven secretion of more IL-6, followed by association of the IL-6, IL-6R complex with EGFR, through direct or indirect interactions, and JAK-dependent phosphorylation of Y1068 of EGFR, leading to EGFR-dependent activation of STAT3, which is immune to inhibition by SOCS3. Because ERK activation in response to EGF is much stronger than ERK activation in response to IL-6, we conclude that EGFR is not completely activated in the second wave, probably because not all of the relevant tyrosine residues can be phosphorylated by the JAKs that are likely to be brought to EGFR in the complex with IL-6R. Many details of this unique receptor–receptor interaction remain to be elucidated in the future. For example, which tyrosine residues of EGFR are susceptible to cross-phosphorylation by the JAKs and which are not, what is the stoichiometry of the receptor–receptor complex and are other membrane-bound proteins involved, do additional EGFR-associated proteins play a role in the second wave, and how does SOCS3 interact with the receptor–receptor complex?
Fig. 4.
Model for the two-phase activation of STAT3 in response to IL-6.
The second wave helps explain why cells with high levels of EGFR have prolonged and enhanced IL-6-dependent activation of STAT3. Aberrant regulation of STAT3 phosphorylation is well known to contribute importantly to diseases involving altered regulation of immunity or cell proliferation. Our expression analysis suggests that the biphasic pattern prolongs and induces the expression of a subset of IL-6-dependent genes, many of which are involved in metabolism and immune regulation (Table 1). All the listed genes are induced in both the initial IL-6 response and in the second wave. In our array data, IRF1 and IRF8, well-characterized immune regulatory proteins, are highly induced in the second wave. Their expression is essential for the IFN response and for immune cell development and differentiation (29, 30). MAPK8 (also known as JNK) has been suggested to play a key role in T-cell proliferation, apoptosis, and differentiation. PRKAA1 (also known as AMPK) regulates the activities of a number of key metabolic enzymes through phosphorylation. It protects cells from stresses that cause ATP depletion and also is important for innate and adaptive immune responses (31, 32).
In cancer cells, aberrant IL-6 signaling and constitutive activation of STAT3 are closely related to cell proliferation and drug resistance. We summarize in Table 1 the enhanced expression, resulting from the second wave of STAT3 activation, of several proteins that play important roles in cancer and inflammation. FOS, a proto-oncogene that is regulated by STAT3, is overexpressed in a variety of human cancers and plays a vital role in the epithelial–mesenchymal transition and aberrant metastatic growth (33, 34). MDR1, a well-known ATP-dependent drug efflux pump, is responsible for decreased drug accumulation in multidrug-resistant cells and often mediates resistance to drugs in lung and breast cancer (35–37). Inhibition of STAT3 can reverse drug resistance in leukemia (38). It has been reported that inhibiting IL-6 and EGFR signaling simultaneously results in a more effective way to control non–small-cell-lung-cancer proliferation, and inhibition of STAT3 activity can increase the sensitivity to tyrosine kinase inhibitors in head and neck cancer (39). Because we have shown here that high expression of EGFR in cancer cells prolongs STAT3 activation in response to IL-6 and OSM (and possibly in response to other cytokines that use the common gp130 receptor), combining inhibitors of EGFR kinase activity, such as erlotinib, with inhibitors of activated STAT3 may have synergistic activity in several different cancers.
In summary, we have found a unique mechanism that explains the biphasic pattern of STAT3 activation in response to IL-6 treatment of cells in which EGFR participates in a positive feedback loop that enhances immune responses and cell proliferation by preventing inhibition by the potent negative regulator SOCS3. The problem of how to sustain the phosphorylation of STAT3 when it is needed during a long period, for example, during normal development, may well be addressed by more than one mechanism. For example, SOCS3 synthesis may be prevented, the protein may be degraded, or it may be inactivated by posttranslational modification. One possibility, as we show here, is that STAT3 phosphorylation can be sustained in the presence of functional SOCS3 if it is catalyzed by a receptor that is not inhibited by SOCS3, and receptors other than EGFR may well play such a role in normal processes. However, in the context of cancer, in which constitutive activation of STAT3 makes a major contribution to the ability of cancer cells to survive and grow, the simultaneous upregulation of EGFR and secretion of IL-6 can cooperate to desensitize the cancer cells to the actions of negative regulators, especially SOCS3.
Materials and Methods
Cells and Reagents.
The human colon carcinoma cell line DLD1, HEK293T cells, the human liver hepatocellular carcinoma cell line HepG2, and the human fibrosarcoma cell line 2fTGH were obtained from the American Type Culture Collection. The cells were grown in DMEM (Gibco-BRL), supplemented with 5% (wt/vol) FBS with penicillin (100 U/mL), and streptomycin (100 μg/mL). AZD1480 was from selleckchem. Antibodies against total STAT3, Y705-phosphoryl STAT3 (pY705-STAT3), and SOCS3 were from Cell Signaling Technology; antibodies against GAPDH and IL-6R were from Santa Cruz, and antibodies against total EGFR were from Bethyl Laboratories. Human recombinant IL-6, IL-6-soluble receptor, and anti-human IL-6 were from Peprotech.
Western Analysis.
Cells were suspended in lysis buffer (50 mM Tris⋅HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing a mixture of protease and phosphatase inhibitors (Roche). After incubation on ice for 20 min, cell debris was removed by centrifugation. Protein (30 μg) was loaded onto 8% (wt/vol) SDS/PAGE gels. The separated proteins were transferred to PVDF membranes (Millipore). The membranes were incubated with primary antibody for 2 h, followed by incubation with secondary antibody for 1 h at room temperature.
Immunoprecipitation and Native Membrane Protein Extraction.
Cells from 100-mm dishes, either treated with IL-6 (100 ng/mL) for 1.5, 2.5, or 4.5 h or untreated, were collected and lysed by using the ProteoExtract Native Membrane Protein Extraction Kit (Calbiochem) according to the manufacturer’s instructions. For immunoprecipitation of EGFR, Protein A Sepharose CL-4B (GE Healthcare Life Sciences) was used. The supernatant solution prepared by using the kit (500 μg enriched membrane-bound protein) was incubated with 2 μg total EGFR antibody overnight at 4 °C and then incubated with prepared Protein A Sepharose beads for 3 h at 4 °C. The immune complexes were recovered by low-speed centrifugation, and the beads were washed with TBS (50 mM Tris⋅HCl at pH 7.4, 150 mM NaCl). The immunoprecipitated proteins were eluted by boiling the beads in the loading buffer for western analysis.
Cytokine Protein Array and ELISA.
DLD1 cells were treated with IL-6 (100 ng/mL) for 30 min, followed by washing with PBS and replacement of the media with or without cycloheximide (CHX, 1 μg/mL). The conditioned media were harvested and analyzed, using the Human Cytokine Array, Panel A (R&D Systems) or the Human IL-6 Quantikine ELISA kit (R&D Systems), following instructions provided by the manufacturer.
Real-Time PCR.
cDNA was synthesized from total RNA, using a modified manufacturer’s protocol with random hexamer and SuperScript III (Invitrogen). Real-time PCR was performed with SYBR Green qPCR master mix (USB) in an iCycler iQ real-time PCR detection system (Bio-Rad). The PCR protocol was initial activation at 95 °C for 5 min, 40 cycles at 95 °C for 15 s, and at 60 °C for 1 min. Aliquots of standard cDNA were included in each PCR, and standard curves for each gene were generated by linear regression. Ct values were converted to gene expression levels by using standard curves. Each gene expression value was normalized to the expression level of GAPDH. Specificity was confirmed by analysis of the melting curves of the PCR products.
Constructs and Gene Transfection.
Vectors (pCDNA3.1) expressing human wild-type EGFR or the EGFR mutants Y992/1173F, Y1045F, Y1068F, and Y1086F were kindly provided by Mark Frey (Children's Hospital, Los Angeles, CA). Each construct was transfected into 293T cells using Lipofectamine Plus (Invitrogen), according to the instructions in the manual. shRNAs for EGFR were purchased from Sigma-Aldrich. shRNA lentiviral transduction was performed according to instructions from the manufacturer.
Gene Expression Analysis.
Total RNA was isolated by using a Qiagen RNeasy Mini Kit according the manufacturer’s instructions, and 1 μg of this RNA was used for microarray analysis on an Illumina HumanHT-12 v4 Expression BeadChip Kit. Data were analyzed by using the Illumina BeadStudio software and normalized by the quantile method. Genes were selected that satisfied the following criteria: differential P values ≤ 0.01, average signals >30, and signals that changed by >1.5-fold.
Supplementary Material
Acknowledgments
We thank Dr. Mark Frey (Children's Hospital, Los Angeles) for generously providing vectors expressing human wild-type or mutant EGFR. This work was supported by National Institutes of Health Grant P01CA062220 (to G.R.S.), Ministry of Science and Technology of the People’s Republic of China Grant 2009DFA30990, Gansu Provincial Ministry of Science and Technology Grant 0708WCGA149, and National Natural Science Foundation of China Grant 2009AA01A130 (to J.Y.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315862110/-/DCSupplemental.
References
- 1.Heinrich PC, et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Croker BA, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4(6):540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 4.Xia L, et al. Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J Biol Chem. 2002;277(34):30716–30723. doi: 10.1074/jbc.M202823200. [DOI] [PubMed] [Google Scholar]
- 5.Kershaw NJ, et al. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. 2013;20(4):469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: Biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318(2):124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leaman DW, et al. Roles of JAKs in activation of STATs and stimulation of c-fos gene expression by epidermal growth factor. Mol Cell Biol. 1996;16(1):369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downward J, Parker P, Waterfield MD. Autophosphorylation sites on the epidermal growth factor receptor. Nature. 1984;311(5985):483–485. doi: 10.1038/311483a0. [DOI] [PubMed] [Google Scholar]
- 9. Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Bio 1(1) [DOI] [PMC free article] [PubMed]
- 10.Gerwin N, Jia GQ, Kulbacki R, Gutierrez-Ramos JC. Interleukin gene expression in mouse preimplantation development. Dev Immunol. 1995;4(3):169–179. doi: 10.1155/1995/26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do DV, et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27(12):1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatt H, Brunet LJ, Stewart CL. Uterine expression of leukemia inhibitory factor coincides with the onset of blastocyst implantation. Proc Natl Acad Sci USA. 1991;88(24):11408–11412. doi: 10.1073/pnas.88.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen MM, Leder P. Leukemia inhibitory factor is expressed by the preimplantation uterus and selectively blocks primitive ectoderm formation in vitro. Proc Natl Acad Sci USA. 1992;89(17):8240–8244. doi: 10.1073/pnas.89.17.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart CL, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359(6390):76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 15.Kopf M, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368(6469):339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AW, et al. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc Natl Acad Sci USA. 2001;98(16):9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945–954. [PubMed] [Google Scholar]
- 18.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13(4-5):357–368. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 21.Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: Implications for translational therapeutics. Cancer. 2007;110(9):1911–1928. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 22.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8(12):1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 23.Leu CM, Wong FH, Chang C, Huang SF, Hu CP. Interleukin-6 acts as an antiapoptotic factor in human esophageal carcinoma cells through the activation of both STAT3 and mitogen-activated protein kinase pathways. Oncogene. 2003;22(49):7809–7818. doi: 10.1038/sj.onc.1207084. [DOI] [PubMed] [Google Scholar]
- 24.Alberti C, et al. Ligand-dependent EGFR activation induces the co-expression of IL-6 and PAI-1 via the NFkB pathway in advanced-stage epithelial ovarian cancer. Oncogene. 2012;31(37):4139–4149. doi: 10.1038/onc.2011.572. [DOI] [PubMed] [Google Scholar]
- 25.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117(12):3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormald S, et al. The comparative roles of suppressor of cytokine signaling-1 and -3 in the inhibition and desensitization of cytokine signaling. J Biol Chem. 2006;281(16):11135–11143. doi: 10.1074/jbc.M509595200. [DOI] [PubMed] [Google Scholar]
- 27.Shao H, Cheng HY, Cook RG, Tweardy DJ. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63(14):3923–3930. [PubMed] [Google Scholar]
- 28.Barnes TC, Anderson ME, Moots RJ. The many faces of interleukin-6: The role of IL-6 in inflammation, vasculopathy, and fibrosis in systemic sclerosis. Int J Rheumatol. 2011;2011:721608. doi: 10.1155/2011/721608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 30.Kano S, et al. The contribution of transcription factor IRF1 to the interferon-gamma-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat Immunol. 2008;9(1):34–41. doi: 10.1038/ni1538. [DOI] [PubMed] [Google Scholar]
- 31.Mayer A, Denanglaire S, Viollet B, Leo O, Andris F. AMP-activated protein kinase regulates lymphocyte responses to metabolic stress but is largely dispensable for immune cell development and function. Eur J Immunol. 2008;38(4):948–956. doi: 10.1002/eji.200738045. [DOI] [PubMed] [Google Scholar]
- 32.Bai A, et al. AMPK agonist downregulates innate and adaptive immune responses in TNBS-induced murine acute and relapsing colitis. Biochem Pharmacol. 2010;80(11):1708–1717. doi: 10.1016/j.bcp.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Mahner S, et al. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer. 2008;99(8):1269–1275. doi: 10.1038/sj.bjc.6604650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Sun Y, Jiang M, Zhang S, Wolfl S. FOS proliferating network construction in early colorectal cancer (CRC) based on integrative significant function cluster and inferring analysis. Cancer Invest. 2009;27(8):816–824. doi: 10.1080/07357900802672753. [DOI] [PubMed] [Google Scholar]
- 35.Leith CP, et al. Correlation of multidrug resistance (MDR1) protein expression with functional dye/drug efflux in acute myeloid leukemia by multiparameter flow cytometry: Identification of discordant MDR-/efflux+ and MDR1+/efflux- cases. Blood. 1995;86(6):2329–2342. [PubMed] [Google Scholar]
- 36.Nakagawa M, et al. Reduced intracellular drug accumulation in the absence of P-glycoprotein (mdr1) overexpression in mitoxantrone-resistant human MCF-7 breast cancer cells. Cancer Res. 1992;52(22):6175–6181. [PubMed] [Google Scholar]
- 37.Amiri-Kordestani L, Basseville A, Kurdziel K, Fojo AT, Bates SE. Targeting MDR in breast and lung cancer: discriminating its potential importance from the failure of drug resistance reversal studies. Drug Resist Updat. 2012;15(1-2):50–61. doi: 10.1016/j.drup.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Xiao W, Wang L, Tian Z, Zhang J. Deactivation of signal transducer and activator of transcription 3 reverses chemotherapeutics resistance of leukemia cells via down-regulating P-gp. PLoS ONE. 2011;6(6):e20965. doi: 10.1371/journal.pone.0020965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen M, et al. Targeting Stat3 abrogates EGFR inhibitor resistance in cancer. Clin Cancer Res. 2012;18(18):4986–4996. doi: 10.1158/1078-0432.CCR-12-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.