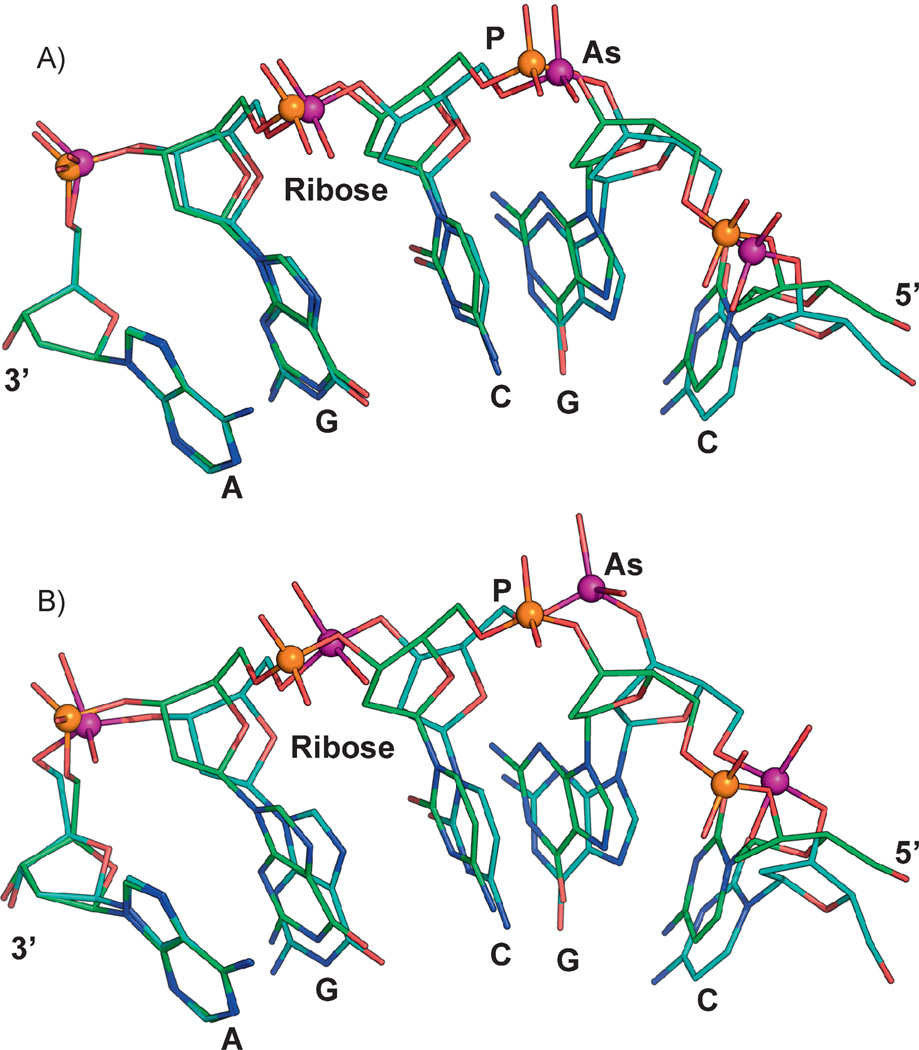
Figure 2.
Molecular modeling of arsenate DNA. The structure of a B-DNA dodecamer (PDB id:1BNA) was used for constructing molecular models. The final model was subject to energy minimization using CHARMM [55]. Initial energy minimization was done with a steep descent algorithm (100 steps) and followed by an adopted basis Newton-Raphson method (1,000 steps). To construct models with arsenate replacing phosphate, the phosphorus atoms in 1BNA were modified to arsenic atoms. Similar changes were incorporated into the CHARMM toplogy and parameter file. In the CHARMM nucleic acid parameter file, the initial O–As distances and O–P–O angles were modified to either (A) 1.7 Å and 109.0° or (B) 1.957 Å and 100.1°. Energy minimization was carried out as described above. Other parameters such as torsion angles and force fields were not modified for these examples. A single strand with five repeating nucleotide units (5′-CGCGA-3’) is shown for both phosphate- (orange sphere) or arsenate- (purple sphere) containing molecules. The phosphate and arsenate models were superimposed to illustrate the difference between the two 5-mers, which are anchored by the adenine base at the 3’ end (left).