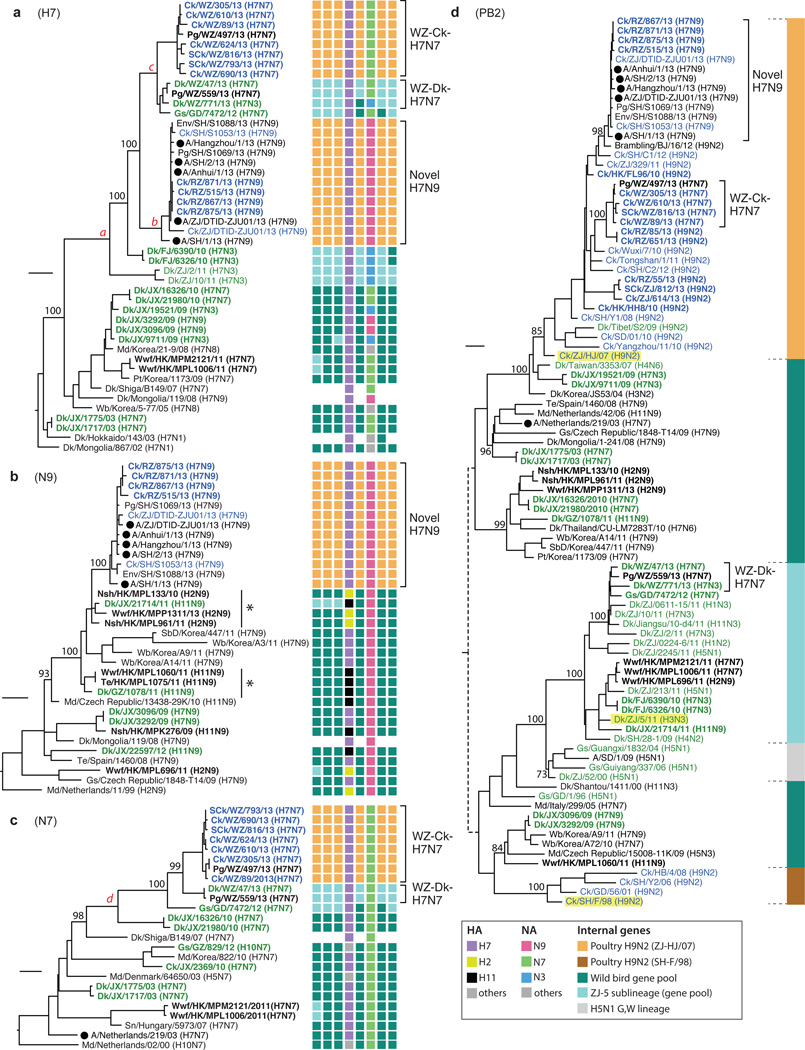
Figure 1. Phylogenies of haemagglutinin, neuraminidase and PB2 genes.
Phylogenies of a, H7 haemagglutinin (n=46); b, N9 neuraminidase (n=34); c, N7 neuraminidase (n=25); d, PB2 (n=93) genes. Sequences reported in this study have their taxa names shown in bold. Genotypes of the influenza viruses are shown on the right (a, b, c) as eight coloured blocks representing each gene segment (from left to right: PB2, PB1, PA, HA, NP, NA, M, and NS; absent if the sequence is unavailable) with the colour indicating the subtype (for HA, NA) or lineage (internal genes; indicated by the solid vertical line in panel d) of that segment. Bootstrap support values (%) from 1,000 pseudo-replicates are shown for selected lineages. Support values for lineages ‘a’ – ‘d’ were all 100%. The scale bar to the left of each tree represents 0.01 substitutions/site. The ‘*’ in panel b denotes N9 sub-lineages linking the viruses of domestic ducks and wild birds. Host species are: Ck (chicken), SCk (silkie chicken), Dk (duck), SbD (spot-billed duck), Md (mallard), Gs (goose), Nsh (Northern shoveler), Wb (wild bird), Wwf (wild waterfowl), Te (common teal), Pg (pigeon), Pt (pintail). Geographic locations: SD (Shandong), ZJ (Zhejiang), JX (Jiangxi), GZ (Guizhou), GD (Guangdong), FJ (Fujian), HK (Hong Kong), WZ (Wenzhou), RZ (Rizhao) and SH (Shanghai). Viruses from different hosts are indicated by: humans, circles; chickens, blue names; domestic ducks or geese, green names.